Tetrapyrroles are involved in various functions critical to whole organisms’ viability, including light absorption, electron transfer, and oxygen binding. GUN1 contributes to important biological processes, including plastid protein homeostasis, through transcription, translation, and protein import.
- plastid
- tetrapyrrole
- heme
- chloroplast
- retrograde signaling
- gun mutants
1. Introduction
Tetrapyrroles are involved in various functions critical to whole organisms’ viability, including light absorption, electron transfer, and oxygen binding [1,2][1][2]. Thus, they are essential components of primary metabolism, such as respiration and photosynthesis. Tetrapyrroles contain four pyrroles, aromatic rings containing four carbon atoms and one nitrogen atom, in linear (e.g., bilins) or cyclic (e.g., porphyrins) chemical structures. Porphyrins often chelate central metal ions, such as Co2+, Fe2+ or Fe3+, or Mg2+ ions. Meanwhile, fully conjugated (pigmented) porphyrin rings possess photodynamic properties: they can generate reactive oxygen species (ROS), primarily singlet oxygen under light excitation, which cause photooxidative damage and cell death [3]. Therefore, organisms must strictly regulate tetrapyrrole biosynthesis. In plants and algae, tetrapyrroles’ main end products are siroheme, heme, phytochromobilin, chlorophyll (Chl) a, and Chl b. Although they are synthesized in plastids, these tetrapyrroles are widely distributed, with the exception of Chls. Especially, heme is found throughout the cell. In addition to prosthetic groups’ function, tetrapyrroles have been proposed as signaling molecules that control transcription and intracellular signaling.
2. Biosynthesis of Tetrapyrroles in Plants
2.1. The C5 Pathway and the Common Pathway
In plant cells, tetrapyrrole biosynthesis takes place entirely in the plastid (Figure 1). The first committed precursor for all tetrapyrroles is 5-aminolevulinic acid (ALA). In plants, algae, and many bacteria, ALA is synthesized from glutamate via the C5 pathway [7][4]. In this pathway, glutamate is first ligated with plastid-encoded tRNAGlu to form glutamyl-tRNAGlu, a substrate for plastid protein biosynthesis. The following two enzymes, glutamyl-tRNA reductase (GluTR) and glutamate 1-semialdehyde aminotransferase (GSAT), synthesize ALA from glutamyl-tRNAGlu. In particular, the step of GluTR is the rate-limiting step of total tetrapyrrole biosynthesis, the activity of which is controlled by transcriptional and post-transcriptional regulations [8,9][5][6]. Arabidopsis has two paralogous genes for GluTR, HEMA1 and HEMA2. HEMA1 is light-responsive, is actively expressed in green tissues, and contributes predominantly to Chl biosynthesis [10,11][7][8]. ALA dehydratase condenses two molecules of ALA onto the monopyrrole to form porphobilinogen (PBG). PBG deaminase assembles four PBG molecules, which are further assembled onto the tetrapyrrole precursor uroporphyrinogen III (Urogen III). Three stepwise oxidation steps oxidatively decarboxylate Urogen III and the final step enzyme, protoporphyrinogen IX oxidase, oxidizes the colorless protoporphyrinogen IX to fully conjugated and pigmented protoporphyrin IX (Proto). Alternatively, Urogen III can be directed to siroheme biosynthesis. Since the pathway from ALA to Proto is conserved among most organisms, this pathway is called ‘the common pathway’.
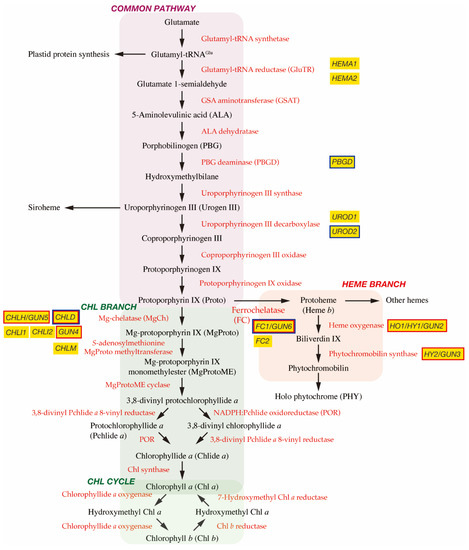
Figure 1. Tetrapyrrole biosynthetic pathway. Enzymes involved in the tetrapyrrole biosynthetic pathway are indicated by red. Important genes described in this article are shown in yellow boxes and genes encoding GUN proteins are indicated by red borders. It is noted that GUN2~GUN6 genes are found at the branch points of Chl and heme biosynthesis. GUN1 interacting proteins are indicated by blue borders.
2.2. Chl Branch and Chl Cycle
The next branchpoint involves the insertion of either Mg2+ or Fe2+ by Mg-chelatase (MgCh) or ferrochelatase (FC), respectively, directing Proto into the Chl or heme biosynthetic pathways. MgCh consists of three subunits, CHLI, CHLD, and CHLH in plants. In Arabidopsis, CHLD and CHLH are encoded by a single gene, and CHLI is encoded by two isoforms, CHLI1 and CHLI2. CHLI1 is essential for photosynthesis [12[9][10],13], whereas CHLI2 has a minor role in assembling the MgCh complex [14][11]. Additionally, GUN4 enhances MgCh activity by mediating substrate or product channeling [15,16,17][12][13][14]. In the Chl branch, MgCh catalyzes the formation of Mg-protoporphyrin IX (MgProto), which is methylated by S-adenosylmethionine MgProto methyltransferase (encoded by CHLM) to form MgProto methyl ester (MgProtoME). MgProtoME cyclase catalyzes the formation of the fifth ring of the tetrapyrrole ring structure, which is further converted to 3,8-divinyl protochlorophyllide a (DV-Pchlide a). DV-Pchlide a is further converted to Pchlide a by DV-Pchlide a 8-vinyl reductase. Pchlide a accumulates in dark-grown angiosperms because the next enzyme, light-dependent NADPH:Pchlide oxidoreductase (POR), requires light to reduce Pchlide a to chlorophyllide a (Chlide a). Depending on the plant species, it is considered that the step of DV-Pchlide a 8-vinyl reductase occurs after POR reaction (Figure 1) [9][6]. Then, Chlide a is esterified with a geranylgeraniol or phytol by Chl synthase to form Chl a, some of which is reversibly converted to Chl b via the Chl cycle [8][5].
2.3. Heme Branch
In the heme branch, FC inserts Fe2+ into Proto to produce protoheme (heme b), which is the prosthetic group of b-type cytochromes and proteins, such as catalase and peroxidase. In these hemoproteins, the heme is noncovalently bound via coordination to the Fe atom by histidine and/or cysteine residues [18][15]. There are two isoforms of FC (FC1 and FC2) in Arabidopsis and cucumber, which show differential tissue-specific and development-dependent expression profiles: FC2 is light-dependent and mainly expressed in photosynthetic tissues, whereas FC1 is stress responsive and ubiquitously expressed in all tissues [19,20][16][17]. Some protoheme is further metabolized into other hemes, such as heme a and heme c. Protoheme is also substrate for bilins. Heme oxygenase oxidatively cleaves protoheme to biliverdin IX. Then, phytochromobilin synthase converts biliverdin IX to 3Z-phytochromobilin. Finally, 3Z-phytochromobilin is isomerized to 3E-phytochromobilin, which functions as the chromophore for the phytochromes (PHYs) [21][18].
3. Coupling of Two Genomes Is Required for Chloroplast Biogenesis
In plant cells, the chloroplast is one of the differentiated states in which plastids have a photosynthetic function [22][19]. In the meristem of angiosperms, plastids exist as undifferentiated proplastids, and chloroplasts can directly form from the proplastids with developmental cues and light signals. This process is called chloroplast biogenesis. During chloroplast biogenesis, thylakoids are formed and stacked into defined grana. The thylakoids are the internal lipid membranes interlaced with protein complexes, which provide the platform for the light reactions of photosynthesis [22,23][19][20]. In the absence of light, proplastids differentiate into etioplasts with unique lattice membrane structures called prolamellar bodies (PLBs), which accumulate Pchlide a and POR. Once the etiolated seedlings are exposed to light, most Pchlide a molecules in PLBs are immediately converted to Chlide a by POR, and then to Chl a via enzymatic processes [24][21].
Plastids originate from a free-living cyanobacterium in a process known as endosymbiosis [25][22]. A primitive cyanobacterium was engulfed by a non-photosynthetic eukaryotic cell and coexisted in ancient times. Many genes of the cyanobacterium endosymbiont are thought to be lost or transferred to the nucleus of the host cell following endosymbiosis. Despite this, some genes involved in photosynthesis, transcription, and translation were retained in plastid genomic DNA [26,27,28][23][24][25]. Photosynthesis in chloroplasts is a reaction that uses light in the photochemical system at the level of thylakoids. The carbon fixation system (Calvin cycle) present in the soluble stroma fraction. Since the protein complexes responsible for these two reaction systems are composed of proteins encoded by nuclear and chloroplast genes, coordinated gene expression between the two components is necessary for functional chloroplast biogenesis.
Thus, for efficient chloroplast biogenesis, communication between the nucleus and the plastids is paramount. The nucleus controls most aspects of chloroplast biogenesis (“anterograde signaling”) [29[26][27],30], while plastids are also thought to emit signals that alter nuclear gene expression (“retrograde signaling”). So far, multiple signaling pathways have been proposed to be plastid-to-nucleus communication. In general, the retrograde signals are categorized into two classes: (i) “biogenic control” signals that mainly act during the initial stage of chloroplast development, and (ii) “operational control” signals that are primarily generated in response to environmental stimuli in matured chloroplasts [31][28]. For evaluation of the biogenic control, the relationship between chloroplast function and nuclear gene expression at the initial stage of seedling development has been mainly evaluated. Meanwhile, the operational control is occurred in matured chloroplasts. This control is proposed to include three chloroplast redox signals: (i) the redox states of components of the photosynthetic electron transport (PET) chain, primarily plastoquinone, (ii) redox-active thiol group-containing proteins and antioxidants couples to PET, and (iii) the generation of ROS [31][28]. As this review focuses the biogenic control, so interested readers are encouraged to see several comprehensive reviews about the operational control [32,33,34][29][30][31].
Important insights into biogenic control of retrograde signals have come from the finding that the expression of many photosynthesis-associated nuclear genes (PhANGs) is dependent on the presence of functional chloroplasts [35,36][32][33]. The perturbation of chloroplast function by mutations or treatments with inhibitors results in the strong down-regulation of many PhANGs [37][34]. Subsequently, a set of mutants, called genomes uncoupled (gun) mutants, which have a reduced ability to coordinate this nuclear response to the chloroplast function, were identified through the retention of PhANGs, such as Lhcb gene expression after treatment with norflurazon (NF) [37][34]. So far, two major categories of mutants have been identified: mutants affected in tetrapyrrole metabolism [12][34][35][36] [15,37,38,39] and mutants in the light signaling components [40][37].
4. Identification of Gun Mutants
The original gun mutant screening isolated five mutants (gun1 to gun5) that retained the expression of PhANGs after NF treatment [37][34]. gun2, gun3, gun4, and gun5 are the four mutants of tetrapyrrole biosynthetic genes and encode heme oxygenase, phytochromobilin synthase, and the regulator and the CHLH subunit of MgCh, respectively [35] [38] (Figure 1). These results suggest the involvement of tetrapyrrole metabolism in biogenic retrograde signaling.
As discussed below, researchers of the retrograde signaling field related to gun mutants have struggled with some of the proposed signals and components of the signaling pathway. These discrepancies may be caused by phenotypic analysis of mutants and transgenic lines involved in retrograde signaling mainly via molecular genetic approaches. These approaches were: knockout or knockdown mutants or transgenic lines of Arabidopsis seedlings, developmental and growth (light intensity and sugar concentration) conditions, type and concentration of inhibitors used, and sensitivity of detection methods (RNA gel blot or quantitative reverse transcription-polymerase chain reaction (qRT-PCR)) in the gun phenotype evaluation (derepression of PhANG expression). In gun mutant screening, NF, an inhibitor of the carotenoid biosynthesis enzyme phytoene desaturase, is mainly used to block chloroplast functions that result in the intense repression of many PhANGs. Inhibition of carotenoid biosynthesis by NF may cause photooxidative stress during the conversion of proplastids to chloroplasts [35,36][32][33]. In general, Arabidopsis seedlings are grown on agar plates containing 1–5 µM NF for 4–10 days under illumination (~100 µmol m−2 s−1) for scoring of the gun phenotype. It is assumed that during growth on the NF-containing plates, free Chl or its precursors accumulate without the concomitant accumulation of carotenoids. In such a situation, ROS (singlet oxygen) are generated, which cause the photooxidative block of chloroplast biogenesis [35,36][32][33]. However, it is not conclusive whether singlet oxygen is generated transiently or consistently, or whether this ROS is directly involved in photooxidative bleaching by NF [6,41][38][39]. It is presumed that NF’s inhibitory mechanism on chloroplast biogenesis may be complex, making it difficult to evaluate the phenotype [42,43][40][41]. Light intensity also affects the ability of NF to repress PhANG expression [42][40]. Furthermore, as there is no clear threshold for determining the gun phenotype. It is sometimes difficult to distinguish whether tested lines are real gun mutants or not, and if the changes in PhANG expression are marginal or rare, but significant.
5. The Function of GUN1
Unlike mutant lines gun 2 to 6 related to the tetrapyrrole biosynthetic pathway, gun1 encodes a chloroplast protein containing a pentatricopeptide repeat (PPR) protein with a C-terminal small MutS-related (SMR) domain. Since gun1 can also prevent down-regulation of PhANG expression after treatment with lincomycin (Lin), an inhibitor of plastid translation [73][42], GUN1 has been suggested to act independently of the tetrapyrrole-mediated GUN signaling pathway. Interestingly, gun1 was shown to be hypersensitive to Lin or NF [96,97,98][43][44][45]. Since the PPR [46] [99] and SMR [47] [100] domains are known to be involved in nucleotide-binding, it was first suggested that GUN1 acts as a nucleotide-binding protein involved in plastid gene expression (PGE), plastid DNA metabolism, or DNA repair [73][42]. Subsequent efforts to screen for GUN1-associated partners by co-immunoprecipitation and mass spectrometry analysis identified many proteins rather than nucleotides [101,102,103][48][49][50]. The highly disordered domain at the N-terminus of GUN1 [51] [104] may correspond to an intrinsically disordered region (IDR) [105][52]. The binding of protein partners induces a conversion of this domain to an ordered structure, which allows the same polypeptide sequence to undertake different interactions with different consequences. Nearly 300 different proteins involved in diverse biological processes in chloroplast were immunoprecipitated after crosslinking of GUN1–GFP in Arabidopsis, suggesting the promiscuous nature of the GUN1 protein [101][48]. Although the specificities to GUN1 were not identified, these putative GUN1-associated proteins were involved in transcription [97][44], translation [101[48][53],106], and import [107][54], all of which include homeostasis of chloroplast proteins [55][56][57] [108,109,110] (Figure 2). In addition, enzymes involved in tetrapyrrole biosynthesis have been identified by yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays [101,107][48][54] (Figure 2).
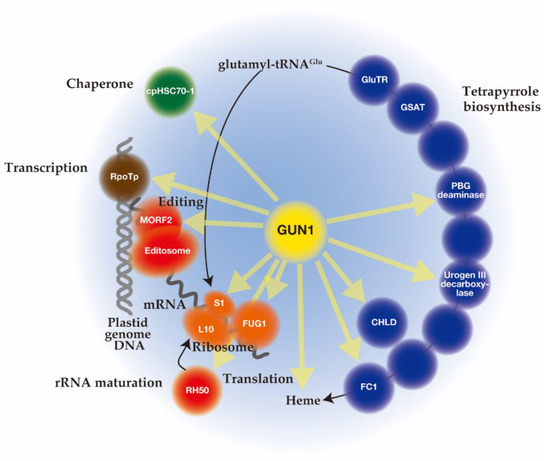
Schematic overview of genomes uncoupled (GUN)1 interacting proteins involved in plastid protein homeostasis (transcription (brown circles), editing and maturation (red circles), translation (orange circles), and chaperone (green circles)) and tetrapyrrole biosynthesis (blue circles). Yellow arrows indicate GUN1 interactions. It is possible that through the N-terminal intrinsically disordered region (IDR) region or pentatricopeptide repeat (PPR) domain, GUN1 forms a droplet that causes molecular crowding, which enhances entropically favor molecular association events, thereby accelerating molecular reactions. Interactions of GUN1 with indicated proteins were demonstrated by co-immunoprecipitation, bimolecular fluorescence complementation (BiFC), and yeast two-hybrid assays.
Although GUN1 is highly and consistently expressed, the protein levels of GUN1 remain not abundant because of its very high turnover [103][50]. The GUN1 protein was only detectable where active chloroplast biogenesis occurs, such as in cotyledons and leaf primordia initially after germination [103][50]. The rapid turnover of GUN1 is controlled mainly by the chaperone ClpC1, suggesting degradation of GUN1 by the Clp protease [103][50]. Inhibition of plastid translation by Lin or oxidative stress by NF may prevent the ClpC-dependent degradation of GUN1, resulting in higher accumulation of this protein under these conditions [103][50]. As GUN1 accumulates only at the very early stage of leaf development under natural conditions, it has been suggested to function in chloroplast biogenesis [103][50]. However, the function of GUN1 at later developmental stages has also been suggested [101,103][48][50]. Since overexpression of GUN1 caused an early flowering phenotype, it is hypothesized that GUN1 functions in developmental phase transitions beyond chloroplast biogenesis [103][50].
Concerning the localization of GUN1 in the plastid, it was first suggested that GUN1 localizes in nucleoids where plastid DNA is actively transcribed. Transiently expressed GUN1–GFP in tobacco (Nicotiana benthamiana) exhibited granular fluorescence colocalizing with pTAC2, a component of transcriptionally-active complexes [73][42]. Such fluorescence in GUN1 was also observed in the stable Arabidopsis GUN1–GFP line [48] [101] and BiFC assays of GUN1 and its binding proteins [97,101][44][48]. Meanwhile, GUN1–GFP was detected in the stroma as a dispersed signal in the stably transformed Arabidopsis lines [103,107][50][54]. It was recently reported that GUN1 alters its sub-chloroplast localization after NF treatment [111][58]: a speckled pattern of fluorescence was detected in the untreated condition, while a diffused distribution was observed after NF treatment. Therefore, it is likely that such a different distribution of GUN1 may be caused by employed developmental stage or functionality of GUN1.
References
- Battersby, A. Tetrapyrroles: The pigments of life. Nat. Prod. Rep. 2000, 17, 507–526.
- Battersby, A.R.; Fookes, C.J.; Matcham, G.W.; McDonald, E. Biosynthesis of the pigments of life: Formation of the macrocycle. Nature 1980, 285, 17–21.
- Op den Camp, R.G.L.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Göbel, C.; Feussner, I.; et al. Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332.
- Ilag, L.; Kumar, A.; Soll, D. Light regulation of chlorophyll biosynthesis at the level of 5-aminolevulinate formation in Arabidopsis. Plant Cell 1994, 6, 265–275.
- McCormac, A.C.; Fischer, A.; Kumar, A.M.; Soll, D.; Terry, M.J. Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J. 2001, 25, 549–561.
- Koncz, C.; Mayerhofer, R.; Koncz-Kalman, Z.; Nawrath, C.; Reiss, B.; Redei, G.; Schell, J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990, 9, 1337–1346.
- Rissler, H.M.; Collakova, E.; DellaPenna, D.; Whelan, J.; Pogson, B.J. Chlorophyll biosynthesis. Expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol. 2002, 128, 770–779.
- Kobayashi, K.; Mochizuki, N.; Yoshimura, N.; Motohashi, K.; Hisabori, T.; Masuda, T. Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochem. Photobiol. Sci. 2008, 7, 1188–1195.
- Larkin, R.; Alonso, J.; Ecker, J.; Chory, J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 2003, 299, 902–906.
- Adhikari, N.D.; Froehlich, J.E.; Strand, D.D.; Buck, S.M.; Kramer, D.M.; Larkin, R.M. GUN4-Porphyrin Complexes Bind the ChlH/GUN5 Subunit of Mg-Chelatase and Promote Chlorophyll Biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1449–1467.
- Davison, P.; Schubert, H.; Reid, J.; Iorg, C.; Heroux, A.; Hill, C.; Hunter, C. Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry 2005, 44, 7603–7612.
- Schneider, S.; Marles-Wright, J.; Sharp, K.H.; Paoli, M. Diversity and conservation of interactions for binding heme in b-type heme proteins. Nat. Prod. Rep. 2007, 24, 621–630.
- Chow, K.-S.; Singh, D.P.; Walker, A.R.; Smith, A.G. Two different genes encode ferrochelatase in Arabidopsis: Mapping, expression and subcellular targeting of the precursor proteins. Plant J. 1998, 15, 531–541.
- Nagai, S.; Koide, M.; Takahashi, S.; Kikuta, A.; Aono, M.; Sasaki-Sekimoto, Y.; Ohta, H.; Takamiya, K.-i.; Masuda, T. Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol. 2007, 144, 1039–1051.
- Terry, M.J.; Linley, P.J.; Kohchi, T. Making light of it: The role of plant haem oxygenases in phytochrome chromophore synthesis. Biochem. Soc. Trans. 2002, 30, 604–609.
- Pogson, B.J.; Ganguly, D.; Albrecht-Borth, V. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 2015, 1847, 1017–1024.
- Pribil, M.; Labs, M.; Leister, D. Structure and dynamics of thylakoids in land plants. J. Exp. Bot. 2014, 65, 1955–1972.
- Masuda, T.; Takamiya, K.-i. Novel Insights into the Enzymology, Regulation and Physiological Functions of Light-dependent Protochlorophyllide Oxidoreductase in Angiosperms. Photosynth. Res. 2004, 81, 1–29.
- Mereschkowsky, C. Über Natur und Ursprung der Chromatophore im Pflanzenreiche. Biol. Cent. 1905, 25, 593–604.
- Archibald, J.M. The puzzle of plastid evolution. Curr. Biol. 2009, 19, R81–R88.
- Keeling, P.J. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu. Rev. Plant Biol. 2013, 64, 583–607.
- Criscuolo, A.; Gribaldo, S. Large-scale phylogenomic analyses indicate a deep origin of primary plastids within cyanobacteria. Mol. Biol. Evol. 2011, 28, 3019–3032.
- Abdallah, F.; Salamini, F.; Leister, D. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 2000, 5, 141–142.
- Singh, R.; Singh, S.; Parihar, P.; Singh, V.P.; Prasad, S.M. Retrograde signaling between plastid and nucleus: A review. J. Plant Physiol. 2015, 181, 55–66.
- Pogson, B.J.; Woo, N.S.; Forster, B.; Small, I.D. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008, 13, 602–609.
- Nott, A.; Jung, H.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759.
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the Languages of the Chloroplast: Retrograde Signaling and Beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53.
- De Souza, A.; Wang, J.Z.; Dehesh, K. Retrograde Signals: Integrators of Interorganellar Communication and Orchestrators of Plant Development. Annu. Rev. Plant Biol. 2017, 68, 85–108.
- Susek, J.; Chory, J. A tale of two genomes: Role of a chloroplast signal in coordinating nuclear and plastid genome expression. Aust. J. Plant Physiol. 1992, 19, 387–399.
- Gray, J.C.; Sullivan, J.A.; Wang, J.H.; Jerome, C.A.; MacLean, D. Coordination of plastid and nuclear gene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 358, 135–144, discussion 144–135.
- Susek, R.; Ausubel, F.; Chory, J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 1993, 74, 787–799.
- Mochizuki, N.; Brusslan, J.; Larkin, R.; Nagatani, A.; Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 2001, 98, 2053–2058.
- Woodson, J.D.; Perez-Ruiz, J.M.; Chory, J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 2011, 21, 897–903.
- Ruckle, M.E.; DeMarco, S.M.; Larkin, R.M. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 2007, 19, 3944–3960.
- Larkin, R.M. Tetrapyrrole Signaling in Plants. Front. Plant Sci. 2016, 7, 1586.
- Kim, C.; Apel, K. 1O2-mediated and EXECUTER-dependent retrograde plastid-to-nucleus signaling in norflurazon-treated seedlings of Arabidopsis thaliana. Mol. Plant 2013, 6, 1580–1591.
- Voigt, C.; Oster, U.; Börnke, F.; Jahns, P.; Dietz, K.-J.; Leister, D.; Kleine, T. In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signalling. Physiol. Plant 2010, 138, 503–519.
- Tanaka, R.; Kobayashi, K.; Masuda, T. Tetrapyrrole Metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0145.
- Larkin, R.M. Influence of plastids on light signalling and development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130232.
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719.
- Zhao, X.; Huang, J.; Chory, J. Genome uncoupled 1 mutants are hypersensitive to norflurazon and lincomycin. Plant Physiol. 2018, 178.
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719.
- Zhao, X.; Huang, J.; Chory, J. Genome uncoupled 1 mutants are hypersensitive to norflurazon and lincomycin. Plant Physiol. 2018, 178.
- Tadini, L.; Peracchio, C.; Trotta, A.; Colombo, M.; Mancini, I.; Jeran, N.; Costa, A.; Faoro, F.; Marsoni, M.; Vannini, C.; et al. GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J. 2020, 101, 1198–1220.
- Song, L.; Chen, Z.; Larkin, R.M. The genomes uncoupled mutants are more sensitive to norflurazon than wild type. Plant Physiol. 2018, 178.
- Kotera, E.; Tasaka, M.; Shikanai, T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 2005, 433, 326–330.
- Moreira, D.; Philippe, H. Smr: A bacterial and eukaryotic homologue of the C-terminal region of the MutS2 family. Trends Biochem. Sci. 1999, 24, 298–300.
- Tadini, L.; Pesaresi, P.; Kleine, T.; Rossi, F.; Guljamow, A.; Sommer, F.; Mühlhaus, T.; Schroda, M.; Masiero, S.; Pribil, M.; et al. GUN1 Controls Accumulation of the Plastid Ribosomal Protein S1 at the Protein Level and Interacts with Proteins Involved in Plastid Protein Homeostasis. Plant Physiol. 2016, 170, 1817–1830.
- Jia, Y.; Tian, H.; Zhang, S.; Ding, Z.; Ma, C. GUN1-Interacting Proteins Open the Door for Retrograde Signaling. Trends Plant Sci. 2019, 24, 884–887.
- Wu, G.-Z.; Chalvin, C.; Hoelscher, M.; Meyer, E.H.; Wu, X.N.; Bock, R. Control of Retrograde Signaling by Rapid Turnover of GENOMES UNCOUPLED1. Plant Physiol. 2018, 176, 2472–2495.
- Shimizu, T.; Kacprzak, S.M.; Mochizuki, N.; Nagatani, A.; Watanabe, S.; Shimada, T.; Tanaka, K.; Hayashi, Y.; Arai, M.; Leister, D.; et al. The retrograde signaling protein GUN1 regulates tetrapyrrole biosynthesis. Proc. Natl. Acad. Sci. USA 2019, 116, 24900–24906.
- Uversky, V.N. Intrinsically disordered proteins from A to Z. Int. J. Biochem. Cell Biol. 2011, 43, 1090–1103.
- Marino, G.; Naranjo, B.; Wang, J.; Penzler, J.F.; Kleine, T.; Leister, D. Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J. 2019, 99, 521–535.
- Wu, G.Z.; Meyer, E.H.; Richter, A.S.; Schuster, M.; Ling, Q.; Schottler, M.A.; Walther, D.; Zoschke, R.; Grimm, B.; Jarvis, R.P.; et al. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 2019, 5, 525–538.
- Colombo, M.; Tadini, L.; Peracchio, C.; Ferrari, R.; Pesaresi, P. GUN1, a Jack-Of-All-Trades in Chloroplast Protein Homeostasis and Signaling. Front. Plant Sci. 2016, 7, 1449.
- Tadini, L.; Jeran, N.; Pesaresi, P. GUN1 and Plastid RNA Metabolism: Learning from Genetics. Cells 2020, 9, 2307.
- Tadini, L.; Jeran, N.; Peracchio, C.; Masiero, S.; Colombo, M.; Pesaresi, P. The plastid transcription machinery and its coordination with the expression of nuclear genome: Plastid-Encoded Polymerase, Nuclear-Encoded Polymerase and the Genomes Uncoupled 1-mediated retrograde communication. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190399.
- Huang, X.Q.; Wang, L.J.; Kong, M.J.; Huang, N.; Liu, X.Y.; Liang, H.Y.; Zhang, J.X.; Lu, S. At3g53630 encodes a GUN1-interacting protein under norflurazon treatment. Protoplasma 2020, 1–8.
