Leptospirosis is a re-emerging worldwide zoonotic disease. The leptospirosis transmission is linked to maintenance and accidental hosts. In the epidemiology of
Leptospira
some serovar are strictly related to specific maintenance hosts.
- leptospirosis
- Leptospira isolation
- wildlife
1. Introduction
Leptospirosis is a neglected and re-emerging zoonoses caused by a Gram-negative bacterium belonging to the Spirochaetaes phylum, Leptospiraceae family,
Leptospira genus [1,2]. These microorganisms appear spiral-shaped, with a diameter of 0.1 µm and a length of 6–20 µm and a pointed end that is typically folded into a characteristic hook shape [1].
[1][2]. These microorganisms appear spiral-shaped, with a diameter of 0.1 µm and a length of 6–20 µm and a pointed end that is typically folded into a characteristic hook shape [1].
Leptospira
is highly mobile and performs rotational movements around the central axis, translation, undulation, and flexion thanks to two periplasmic axial flagella located under the cell membrane [3]. Although
Leptospira
is a microaerophile, it develops well even in conditions of complete aerobiosis. The optimum temperature for its growth is between 28 °C and 30 °C, although it also grows at 37 °C. The ideal pH range is between 7.2 and 7.4 [2].
Traditional classification divided the genus
Leptospira
in two species:
L. interrogans
, pathogenic strains, and
L. biflexa
, saprophytic strains. On the basis of antigenic agglutination reactions and cross absorption, both species are divided into serovars. There are over 60 serovars belonging to the
L. biflexa
species, while there are more than 200
Leptospira interrogans [1,2]. Serovars are identified on the basis of their expression of surface epitopes in the mosaic of lipopolysaccharide antigens (LPS), where the specificity of the latter depends on the composition and orientation of sugars [4]. Traditionally, antigenic correlated serovars were grouped into serogroups, which are very relevant in the epidemiological field [5]. Currently, the
[1][2]. Serovars are identified on the basis of their expression of surface epitopes in the mosaic of lipopolysaccharide antigens (LPS), where the specificity of the latter depends on the composition and orientation of sugars [4]. Traditionally, antigenic correlated serovars were grouped into serogroups, which are very relevant in the epidemiological field [5]. Currently, the
Leptospira genus has undergone a re-classification on a genomic basis, which resulted in the identification of 13 species in addition to those already existing, reaching a total of 64 identified species [
genus has undergone a re-classification on a genomic basis, which resulted in the identification of 13 species in addition to those already existing, reaching a total of 64 identified species [1]. This classification system may be more complicated than the previous one as, within the same species, pathogenic and non-pathogenic serovars are included, and specific serovars can be found within several species. For this reason, in several laboratories, the old classification is still used, mainly for convenience in the serological diagnosis [
1]. This classification system may be more complicated than the previous one as, within the same species, pathogenic and non-pathogenic serovars are included, and specific serovars can be found within several species. For this reason, in several laboratories, the old classification is still used, mainly for convenience in the serological diagnosis [1]. Serovars, identified on a genomic basis, amount to more than 260 and are classified into pathogenic, intermediate, and saprophytic [6,7]. Pathogenic
]. Serovars, identified on a genomic basis, amount to more than 260 and are classified into pathogenic, intermediate, and saprophytic [6][7]. Pathogenic
Leptospira
are the causative agents of moderate to severe forms of the disease, while intermediate
Leptospira generally cause less severe infections. On the other hand, saprophytic serovars, commonly present in the environment, are not considered pathogenic and can play a relevant role only when they undergo genetic recombination processes with pathogenic serovars [8,9].
generally cause less severe infections. On the other hand, saprophytic serovars, commonly present in the environment, are not considered pathogenic and can play a relevant role only when they undergo genetic recombination processes with pathogenic serovars [8][9].
The invasion of
Leptospira
into the body occurs through skin lesions (even of minimal entity), via the mucous membrane (conjunctiva and oral mucosa), and by contact with wet skin or by inhalation [2]. Infiltrated microorganisms invade the bloodstream, causing bacteremia that persists for about 5–7 days [10]. Once a critical number of bacteria has been reached in the blood, the first symptoms related to their trans-endothelial migration appear. The pathogenetic mechanism of
Leptospira is not yet fully understood, and it is hypothesized that virulence factors, such as toxins, adhesins, and other surface proteins, are expressed [11]. Primary lesions affect the endothelium of small vessels and cause ischemic damage in various organs, including kidneys, liver, lungs, meninges, placenta, and muscles [10]. In certain cases, hemorrhages, jaundice due to the destruction of the hepatic architecture, and, more frequently, thrombocytopenia, may also occur [11]. Tissue damage, although severe, can undergo complete healing or its resolution can leave scar tissue, as is often observed in pig kidneys with the characteristic appearance of “white spots” [2].
is not yet fully understood, and it is hypothesized that virulence factors, such as toxins, adhesins, and other surface proteins, are expressed [11]. Primary lesions affect the endothelium of small vessels and cause ischemic damage in various organs, including kidneys, liver, lungs, meninges, placenta, and muscles[10]. In certain cases, hemorrhages, jaundice due to the destruction of the hepatic architecture, and, more frequently, thrombocytopenia, may also occur [11]. Tissue damage, although severe, can undergo complete healing or its resolution can leave scar tissue, as is often observed in pig kidneys with the characteristic appearance of “white spots” [2].
2.
Leptospira
Isolation on “Unconventional” Host
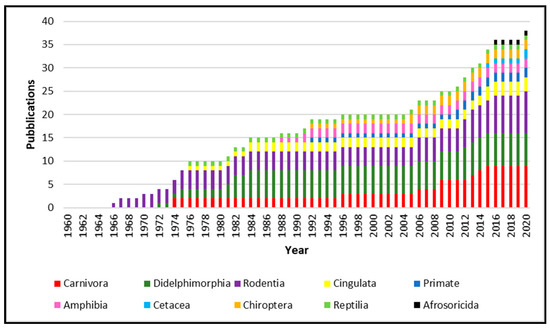
From 1960 to 2020, 34 papers were published about the isolation of
Leptospira
from species not recognized as leptospirosis reservoir. As showed in
, the published works increase constantly year after year, reporting
Leptospira
isolation from several animal species.
Leptospira
spp. isolated from animals belonging to the Carnivora order are the most numerous, followed by Didelphimorphia and Rodentia. However, isolation was obtained from Cetacea, Cingulata, Afrosoricida, Chiroptera and Primate, as well as in Reptilia and Amphibia classes.
The geographical distribution of
Leptospira
isolation (
) is more abundant in South America, especially in Brazil and Argentina, due to the high animal species variability present in this geographic area. Moreover, other isolations were performed in the North American West Coast, Italy, Netherlands, Japan, and Madagascar.
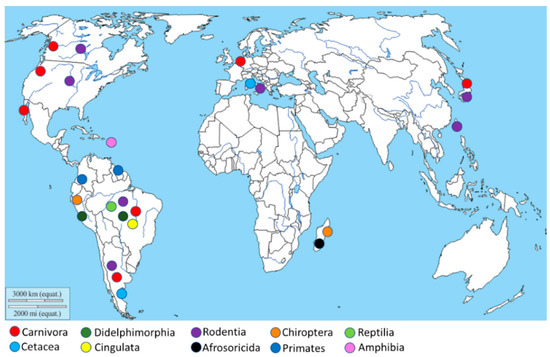
Figure 2.
Geographical distribution of the
Leptospira
isolation. The map shows, through different colors,
Leptospira
isolation from different animal orders.
3. Conclusions
Leptospirosis is probably the most prevalent, underestimated, and re-emerging zoonotic disease; animals, both wild and domestic, represent one of its most important transmission sources.
Leptospira
infection is endemic in many countries with no surveillance or diagnostic facilities, especially for animals, such as in several African countries where leptospirosis has rarely or never been reported. These findings are undoubtedly of high importance for human public health, due to the risk of human infection through interaction with the reservoir or incidental hosts or contact with biological materials, including blood, urine, tissue, and excretions. Such risks could affect not only already confirmed worker categories (i.e., veterinarians, trappers, abattoir workers, farm workers, hunters, animal shelter workers and scientists and technologists handling animals in laboratories or during field work) but also marine mammal workers, fishmen, researchers, wildlife rehabilitators, trainers, and zoological park workers.
A better knowledge of the epidemiology of this infectious disease is essential to facilitate the creation of efficient prevention and control programs using a One Health approach. Constant monitoring is needed to control the evolution of the dynamics of leptospirosis epidemiology, mainly focused on new animal species that could contribute to its spreading, in order to better clarify their role as a reservoir or incidental hosts.
References
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326.
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296.
- Goldstein, S.F.; Charon, N.W. Motility of the spirochete leptospira. Cell Motil. Cytoskelet. 1988, 9, 101–110.
- Adler, B. Leptospira and Leptospirosis. In Current Topics in Microbiology and Immunology; Springer: Berlin, Germany, 2015; ISBN 978-3-662-45059-8.
- Kmety, E.; Dikken, H. Classification of the Species Leptospira Interrogans and History of its Serovars; University Press Groningen: Groningen, The Netherlands, 1993.
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374.
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Mohd Khalid, M.K.N.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270.
- Balamurugan, V.; Gangadhar, N.L.; Mohandoss, N.; Thirumalesh, S.R.A.; Dhar, M.; Shome, R.; Krishnamoorthy, P.; Prabhudas, K.; Rahman, H. Characterization of leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. Springerplus 2013, 2, 362.
- Barragan, V.; Chiriboga, J.; Miller, E.; Olivas, S.; Birdsell, D.; Hepp, C.; Hornstra, H.; Schupp, J.M.; Morales, M.; Gonzalez, M.; et al. High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador. PLoS Negl. Trop. Dis. 2016, 10, e0004990.
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 3, 757–771.
- Levett, P.N.; Haake, D.A. Leptospira Species (Leptospirosis). In Principles and Practice of Infectious Diseases; Churchill Livingstone Elsevier: Philadelphia, PA, USA, 2010; pp. 3059–3065.
