Candida albicans is a common commensal fungus that colonizes the oropharyngeal cavity, gastrointestinal and vaginal tract, and healthy individuals’ skin.
- Candida albicans
- virulence traits
1. Introduction
1.1. Fungal Infection
1.2. Candida Albicans
Candida albicans Pappears in several morpthological forms (blastospores, pseudohyphae, and hyphae) (Figure 1). Blaogenstospores divide asexually by budding [5,6]. During that process, new cell material is formed on the surface of the blastospore. The new bud grows from a small selected blastospore, and it is most often located distally from the site of a scar caused by birth, after which the phase of growth begins. After the growth phase ends, the cells divide, whereby the daughter separates fromauses fungal infections, such as the parent cell by creating a partition [6].
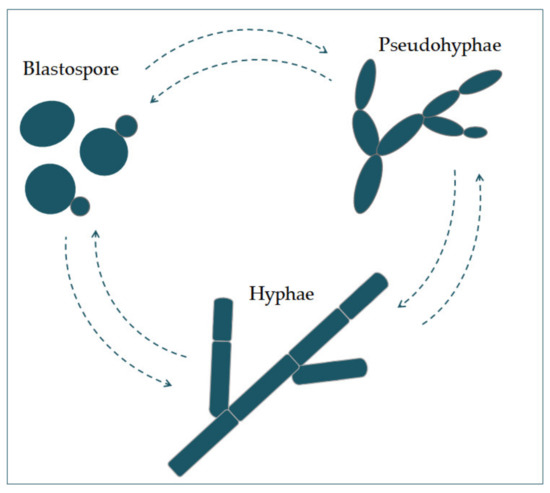
Chains, of elongated yeast cells characterize pseudohyphae, and the shape of hyphae is characterized by branched chains of tubular cells, with no narrowing at the sites of septation [7]. Filre widespread and may affect the skin amentation is enhanced by a temperature higher than 37 °C, an alkali pH, serum, and high concentrations of CO2 [8]. In the sd mucosal surfamce way, it is also enhanced by a lack of nitrogen and carbon in the presence of N-acetylglucosamine (GlcNAc) [7]. Th, and may cause systemic is transition from a blastospore to a hypha is characterized by the activationfection. Species of ofCandida are complex regulatory network of signal paths, which include many transcription factors [8]. Thpresent in as many as 400,000 syste main difference between yeast and hyphae composition is that theic fungal diseases hypha[1]. wallOf has slightly more chitin content than yeast [9].
The cell wall iall the s madpe of glucan, chitin, and protein. Itscies, roleCandida albicans is to protect the cell from stressful conditions in the environment, such as osmotic changes, dehydration, and temperature changes, and protect the cells from the host’s immune defense [10,11]. It is most common causative agent of mucosalso responsible for adhesion to the host cell, with adhesion proteins such as Als1-7, Als9, and Hwp1 [12].
Communication of the cell with the outinfections and side environment takeys place through the cell membrane [13]. Sterols temic in thfe cell membrane are extremely important, giving the cell stability, rigidity, and resistance to physical stressors [9]. Ection, and it is rgosterol is the most represented sterol and is characteristic for the cell membrane sponsible for about 70% of fungi. It is synthesized on the endoplasmic reticulum anal infections around the world lipid bodies [14][2]. In the cell membrane, there is a phospholipid bilayer containing proteins with the role of receptors, but also some whose role is transport and also signal transduction [15].
In its met has been the leading cabolism, Candida albicans uses glucose as a source of carbon and amino acids as nitrogen sources [16].
2. Virulence Factors of Candida albicans
Candida pof life-threarticipates actively in the pathophysiology of the occurrence and advance of ening invasive infection, thanks to its virulence factors. One group of virulence factors causes colonization to take place, or the initiation of an infection, whilst the other group helps to spread the infection [17].
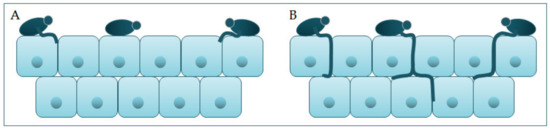
Polymorphism implies s for the past several decades. Despite treatment, the transition of C. albicans from a commensal form to a pathological one, which depends on changes in the environment in which it is located. It is characterized by the morphological transition of blastospores into hyphae, and the transitional form between are pseudohyphae [18,19]. Asexuartality rate is close to 40%, especial spores, chlamydospores, are formed under adverse conditions and are three to four times bigger than blastospores [12]. The my in hospital corphological transition of C. albicans beginditions with[3][4]. tThe budding of blastospores and the formation of new cells. The nuclei separate at the mother–daughter cell junction via the septum [20]. The present review aims to provide anuclei of hyphal forms divide in the germ tube but outside the septum region. After division, one nucleus returns to the mother cell, and the other moves toward the center overview of the virulence traits of the germ tube [21]. C.andida albicans is preseant in the form of yeast in the human microbiome. The transition from yeast to the hyphal form is a transition into a pathogenic form [22,23]. The hyphd its clinical manifestations in the oral form is invcasive, and in this form, the cells enter the host tissue by active penetration and induced endocytosis [24] (Figure 2). Induced endocytosis is medivity, intestinal mucosated, by hyphae invasion and depends on host activity, whereas active penetration depends on the fungal activity [25]. Severskin, as well as in inval signaling pathways are involved in hyphal formation. The most important is cAMP-dependent protein kinase A (cyclic adenosine monophosphate PKA) [3,26]ive infections.
1.2. Candida Albicans
Candida albicans appears in several morphological forms (blastospores, pseudohyphae, and hyphae) (Figure 1). Blastospores divide asexually by budding [5][6]. During that process, new cell material is formed on the surface of the blastospore. The new bud grows from a small selected blastospore, and it is most often located distally from the site of a scar caused by birth, after which the phase of growth begins. After the growth phase ends, the cells divide, whereby the daughter separates from the parent cell by creating a partition [6].

Figure 1. The morphological switches and transitions of Candida albicans during the infection process. The morphological transitions from blastospore to pseudohyphae and hyphae are reversible.
Chains of elongated yeast cells characterize pseudohyphae, and the shape of hyphae is characterized by branched chains of tubular cells, with no narrowing at the sites of septation [7]. Filamentation is enhanced by a temperature higher than 37 °C, an alkali pH, serum, and high concentrations of CO2 [8]. In the same way, it is also enhanced by a lack of nitrogen and carbon in the presence of N-acetylglucosamine (GlcNAc) [7]. This transition from a blastospore to a hypha is characterized by the activation of a complex regulatory network of signal paths, which include many transcription factors [8]. The main difference between yeast and hyphae composition is that the hypha wall has slightly more chitin content than yeast [9].The cell wall is made of glucan, chitin, and protein. Its role is to protect the cell from stressful conditions in the environment, such as osmotic changes, dehydration, and temperature changes, and protect the cells from the host’s immune defense [10][11]. It is also responsible for adhesion to the host cell, with adhesion proteins such as Als1-7, Als9, and Hwp1 [12].Communication of the cell with the outside environment takes place through the cell membrane [13]. Sterols in the cell membrane are extremely important, giving the cell stability, rigidity, and resistance to physical stressors [9]. Ergosterol is the most represented sterol and is characteristic for the cell membrane of fungi. It is synthesized on the endoplasmic reticulum and lipid bodies [14]. In the cell membrane, there is a phospholipid bilayer containing proteins with the role of receptors, but also some whose role is transport and also signal transduction [15].In its metabolism, Candida albicans uses glucose as a source of carbon and amino acids as nitrogen sources [16].
2. Virulence Factors of Candida albicans
Candida participates actively in the pathophysiology of the occurrence and advance of infection, thanks to its virulence factors. One group of virulence factors causes colonization to take place, or the initiation of an infection, whilst the other group helps to spread the infection [17]. Polymorphism implies the transition of C. albicans from a commensal form to a pathological one, which depends on changes in the environment in which it is located. It is characterized by the morphological transition of blastospores into hyphae, and the transitional form between are pseudohyphae [18][19]. Asexual spores, chlamydospores, are formed under adverse conditions and are three to four times bigger than blastospores [12]. The morphological transition of C. albicans begins with the budding of blastospores and the formation of new cells. The nuclei separate at the mother–daughter cell junction via the septum [20]. The nuclei of hyphal forms divide in the germ tube but outside the septum region. After division, one nucleus returns to the mother cell, and the other moves toward the center of the germ tube [21]. C. albicans is present in the form of yeast in the human microbiome. The transition from yeast to the hyphal form is a transition into a pathogenic form [22][23]. The hyphal form is invasive, and in this form, the cells enter the host tissue by active penetration and induced endocytosis [24] (Figure 2). Induced endocytosis is mediated by hyphae invasion and depends on host activity, whereas active penetration depends on the fungal activity [25]. Several signaling pathways are involved in hyphal formation. The most important is cAMP-dependent protein kinase A (cyclic adenosine monophosphate PKA) [3][26].

Schematic presentation of (
) adherence and colonization, and (
) penetration and invasion of
It has been shown that a hypha-specific toxin, candidalysin, is crucial for the occurrence of candidiasis [19,27]. Candidalysin is a cytolytic 31-amino acid α-helical amphipathic peptide [19,28]. It is produced by the
It has been shown that a hypha-specific toxin, candidalysin, is crucial for the occurrence of candidiasis [19][27]. Candidalysin is a cytolytic 31-amino acid α-helical amphipathic peptide [19][28]. It is produced by the
C. albicans hyphae, and it is crucial in damaging the host cells. It is thought that it contributes to establishing a systemic infection and mortality [29]. Candidalysin is capable of directly damaging the epithelial membrane, by intercalation, permeabilization, and creating pores, causing the cytoplasmic contents to weaken [29,30].
Factors that contribute to the pathogenic potential of
hyphae, and it is crucial in damaging the host cells. It is thought that it contributes to establishing a systemic infection and mortality [29]. Candidalysin is capable of directly damaging the epithelial membrane, by intercalation, permeabilization, and creating pores, causing the cytoplasmic contents to weaken [29][30].Factors that contribute to the pathogenic potential of
Candida albicans
are the expression of proteins important for adhesion and invasion. The process of adhesion is affected by various factors, such as the types of protein in the cell wall, and the physical and chemical properties of the cell surface. Adhesins of
C. albicans
recognize ligands such as proteins, fibrinogens, and fibronectins and bind to them [17]. Since adhesins such as Als3 and Hwp1 are mainly expressed during hyphae creation, they play an important role in the adhesion of
C. albicans to the host cells [17].
Formation of biofilm is a property of
to the host cells [17]. Formation of biofilm is a property of
C. albicans
pathogenesis. Most infections caused by
C. albicans
are related to the creation of a biofilm on the surface of the host or on abiotic surfaces (implants), which leads to high morbidity and mortality [23]. Because
C. albicans
can transition from yeast to hyphae morphologically, its biofilm is a complex structure of different morphological forms [31]. The biofilm develops through several consecutive phases [32]. In the first phase, the individual cells of
Candida albicans
adhere to the substrate, which forms the basal layer of the biofilm. After that comes the phase of cell proliferation and filamentation, in which the cells form elongated protrusions, which continue growing into filamentous hyphal forms. The production of hyphae is a sign of the initiation of the creation of the biofilm. In the maturation phase, the accumulation of an extracellular polysaccharide matrix follows. The final phase involves the dispersion of non-adherent cells, which results in the possibility of the inception of new biofilms (
Figure 3) and the possibility of dissemination in the tissue [33,34].
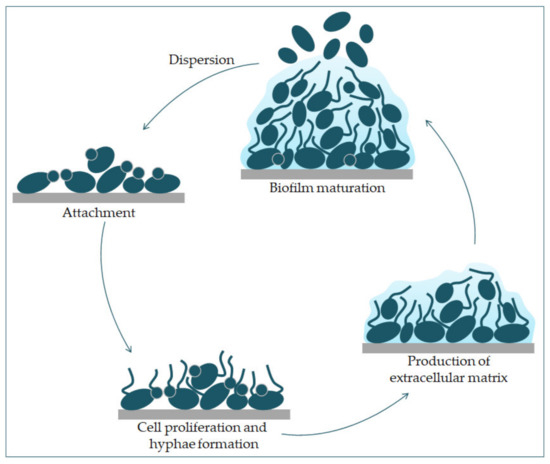
) and the possibility of dissemination in the tissue [33][34].

Phases of
biofilm formation. The formation starts with the attachment of yeast cells (green) to the surface (grey). In the early phase of the biofilm occurs the proliferation of
The extracellular polysaccharide matrix comprises extracellular polymers and extracellular DNA involved in maintaining the biofilm structure [35]. Additionally, extracellular DNA plays a vital role in binding the biofilm to the substrate [32]. An essential part of the extracellular matrix are β-1,3-glucans, which significantly contribute to the biofilm’s resistance to antifungal drugs because they prevent contact with target cells [36].
and hyphal cells’ formation. The production of the extracellular matrix follows. The maturation phase includes the accumulation of an extracellular matrix. Finally, yeast cells disperse to a new site and form a new biofilm.
The extracellular polysaccharide matrix comprises extracellular polymers and extracellular DNA involved in maintaining the biofilm structure [35]. Additionally, extracellular DNA plays a vital role in binding the biofilm to the substrate [32]. An essential part of the extracellular matrix are β-1,3-glucans, which significantly contribute to the biofilm’s resistance to antifungal drugs because they prevent contact with target cells [36].
C. albicans cells in biofilm release more β-1,3-glucans into the extracellular matrix than planktonic cells [37]. The biofilm channels facilitate cell supply with nutrients, air, and water, giving it new “multicellular” properties [32]. Intercellular communication, or quorum sensing, is an essential factor in forming biofilm and is based on microorganisms’ behavior and the synthesis of signal molecules [38]. “Autoinducers” are signal molecules that regulate the population density by a signal mechanism. The binding of signal molecules to receptors suppresses target genes when a specific biofilm density is reached at a critical autoinducers concentration. This modulation of the quorum sensing process maintains the biofilm fungal colony’s optimal size and encodes virulent phenotypes [32]. The transcription network that regulates biofilm formation consists of six major transcription regulators (Efg1, Tec1, Bcr1, Ndt80, Rob1, and Brg1) that regulate the expression of 1000 genes [39,40]. Bcr1 transcription factor (Biofilm and Cell wall Regulator 1), whose main target is Hwp1 (Hyphal Wall Protein), is necessary to form biofilm on mucosal surfaces [41]. The Hwp1 protein binds to transglutaminases on host cells in biofilms on mucosal surfaces. While on abiotic surfaces, it is expressed as an independent enzyme of the host and has an adhesion function [42]. Several different gene products control biofilm development on abiotic surfaces transcription factors (Efg1, Bcr1, Tye7), cell wall proteins (Hwp1, Als3), protein kinases (Ire1, Cbk1) [43]. The two essential regulators of biofilm on abiotic surfaces are Efg1 and Bcr1. These transcription factors are needed for the expression of different genes for cell adhesion and filamentation in biofilms on abiotic surfaces. Additionally, the adhesin Als3 which is the target of Bcr1 plays a crucial role in the formation of biofilm on the abiotic surface [43]. During the formation of a biofilm, besides the change in expression of genes directly involved in its formation, the expression of genes indirectly related to different characteristics of the biofilm also changes [44]. The expression of genes involved in the metabolism of sulfur-containing amino acids is increased, which is characteristic of cells in the biofilm’s deeper layers. This metabolism allows cells to survive starvation and oxidative stress because sulfur amino acids are involved in the synthesis of antioxidants. The biofilm cells form a hypoxic environment and increase the expression of genes involved in glycolysis, fatty acid metabolism, and ergosterol synthesis [45].
Thigmotropism of the hyphae of
cells in biofilm release more β-1,3-glucans into the extracellular matrix than planktonic cells [37]. The biofilm channels facilitate cell supply with nutrients, air, and water, giving it new “multicellular” properties [32]. Intercellular communication, or quorum sensing, is an essential factor in forming biofilm and is based on microorganisms’ behavior and the synthesis of signal molecules [38]. “Autoinducers” are signal molecules that regulate the population density by a signal mechanism. The binding of signal molecules to receptors suppresses target genes when a specific biofilm density is reached at a critical autoinducers concentration. This modulation of the quorum sensing process maintains the biofilm fungal colony’s optimal size and encodes virulent phenotypes [32]. The transcription network that regulates biofilm formation consists of six major transcription regulators (Efg1, Tec1, Bcr1, Ndt80, Rob1, and Brg1) that regulate the expression of 1000 genes [39][40]. Bcr1 transcription factor (Biofilm and Cell wall Regulator 1), whose main target is Hwp1 (Hyphal Wall Protein), is necessary to form biofilm on mucosal surfaces [41]. The Hwp1 protein binds to transglutaminases on host cells in biofilms on mucosal surfaces. While on abiotic surfaces, it is expressed as an independent enzyme of the host and has an adhesion function [42]. Several different gene products control biofilm development on abiotic surfaces transcription factors (Efg1, Bcr1, Tye7), cell wall proteins (Hwp1, Als3), protein kinases (Ire1, Cbk1) [43]. The two essential regulators of biofilm on abiotic surfaces are Efg1 and Bcr1. These transcription factors are needed for the expression of different genes for cell adhesion and filamentation in biofilms on abiotic surfaces. Additionally, the adhesin Als3 which is the target of Bcr1 plays a crucial role in the formation of biofilm on the abiotic surface [43]. During the formation of a biofilm, besides the change in expression of genes directly involved in its formation, the expression of genes indirectly related to different characteristics of the biofilm also changes [44]. The expression of genes involved in the metabolism of sulfur-containing amino acids is increased, which is characteristic of cells in the biofilm’s deeper layers. This metabolism allows cells to survive starvation and oxidative stress because sulfur amino acids are involved in the synthesis of antioxidants. The biofilm cells form a hypoxic environment and increase the expression of genes involved in glycolysis, fatty acid metabolism, and ergosterol synthesis [45].Thigmotropism of the hyphae of
C. albicans
is regulated by the extracellular intake of calcium through calcium channels. It is an important mechanism in the enhancement of the virulence of
Candida spp. Thigmotropism aids in creating a biofilm on abiotic surfaces and the spread in the host tissue [16].
Among virulence factors of
Thigmotropism aids in creating a biofilm on abiotic surfaces and the spread in the host tissue [16]. Among virulence factors of
C. albicans
is phenotype transition between white and opaque cells. Phenotype diversity provides a quick response to changes in the environment. It is extremely important for the life of many microbe species. In
Candida albicans cells, switching between two phenotype states, white and opaque, leads to differences in filamentous growth and interactions with immunological cells in vitro [46]. Morphological changes and phenotypic switches are stabilized transcriptionally and are stable for many generations [47].
Secretion of hydrolytic enzymes are present in
cells, switching between two phenotype states, white and opaque, leads to differences in filamentous growth and interactions with immunological cells in vitro [46] Morphological changes and phenotypic switches are stabilized transcriptionally and are stable for many generations [47].Secretion of hydrolytic enzymes are present in
Candida albicans.
Hydrolytic enzymes facilitate the commensal and pathogenic characteristics such as attachment to host tissue and causing the host cell membrane’s rupture. Because of these enzymes, invasion into the surfaces of mucous membrane and blood vessels is possible, and they also participate in avoiding the host’s immune response. The three main enzymes produced by
C. albicans are SAP (secreted aspartyl protease), phospholipase, and hemolysin [48].
are SAP (secreted aspartyl protease), phospholipase, and hemolysin [48].
References
- Mukaremera, L.; Lee, K.K.; Mora-Montes, H.M.; Gow, N.A.R. Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front. Immunol. 2017, 8, 1.
- Morad, H.O.J.; Wild, A.-M.; Wiehr, S.; Davies, G.; Maurer, A.; Pichler, B.J.; Thornton, C.R. Pre-clinical Imaging of Invasive Candidiasis Using ImmunoPET/MR. Front. Microbiol. 2018, 9, 1996.
- Chen, H.; Zhou, X.; Ren, B.; Cheng, L. The regulation of hyphae growth in Candida albicans. Virulence 2020, 11, 337–348.
- Basmaciyan, L.; Bon, F.; Paradis, T.; Lapaquette, P.; Dalle, F. Candida Albicans Interactions with The Host: Crossing The Intestinal Epithelial Barrier. Tissue Barriers 2019, 7, 1612661.
- Walker, G.M.; White, N.A. Introduction to Fungal Physiology. In Fungi: Biology and Applications; Kavanagh, K., Ed.; Wiley-Blackwell: Chichester, UK, 2017; p. 4. ISBN 978-1-119-37432-9.
- Molero, G.; Díez-Orejas, R.; Navarro-García, F.; Monteoliva, L.; Pla, J.; Gil, C.; Sánchez-Pérez, M.; Nombela, C. Candida albicans: Genetics, dimorphism and pathogenicity. Int. Microbiol. 1998, 1, 95–106.
- Kornitzer, D. Regulation of candida albicans hyphal morphogenesis by endogenous signals. J. Fungi 2019, 5.
- Basso, V.; d’Enfert, C.; Znaidi, S.; Bachellier-Bassi, S. From genes to networks: The regulatory circuitry controlling candida albicans morphogenesis. In Fungal Physiology and Immunopathogenesis: Current Topics in Microbiology and Immunology; Rodrigues, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; Volume 422, pp. 61–99.
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2020, 10, 2993.
- Höfs, S.; Mogavero, S.; Hube, B. Interaction of Candida albicans with host cells: Virulence factors, host defense, escape strategies, and the microbiota. J. Microbiol. 2016, 54, 149–169.
- Reyna-Beltrán, E.; Isaac Bazán Méndez, C.; Iranzo, M.; Mormeneo, S.; Pedro Luna-Arias, J. The Cell Wall of Candida albicans: A Proteomics View. In Candida Albicans; Sandai, D., Ed.; IntechOpen: London, UK, 2019.
- Ciurea, C.N.; Kosovski, I.-B.; Mare, A.D.; Toma, F.; Pintea-Simon, I.A.; Man, A. Candida and Candidiasis—Opportunism Versus Pathogenicity: A Review of the Virulence Traits. Microorganisms 2020, 8, 857.
- Hall, R.A. Dressed to impress: Impact of environmental adaptation on the Candida albicans cell wall. Mol. Microbiol. 2015, 97, 7–17.
- Jordá, T.; Puig, S. Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes 2020, 11, 795.
- Cho, I.; Jackson, M.R.; Swift, J. Roles of Cross-Membrane Transport and Signaling in the Maintenance of Cellular Homeostasis. Cell. Mol. Bioeng. 2016, 9, 234–246.
- Viana, R.; Dias, O.; Lagoa, D.; Galocha, M.; Rocha, I.; Teixeira, M.C. Genome-Scale Metabolic Model of the Human Pathogen Candida albicans: A Promising Platform for Drug Target Prediction. J. Fungi 2020, 6, 171.
- Deorukhkar, S.C. Virulence Traits Contributing to Pathogenicity of Candida Species. J. Microbiol. Exp. 2017, 5.
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 2017, 15, 96–108.
- Hanaoka, M.; Domae, E. IL-1α released from oral epithelial cells upon candidalysin exposure initiates an early innate epithelial response. Int. Immunol. 2020.
- Gale, C.; Gerami-Nejad, M.; McClellan, M.; Vandoninck, S.; Longtine, M.S.; Berman, J. Candida albicans Int1p Interacts with the Septin Ring in Yeast and Hyphal Cells. Mol. Biol. Cell 2001, 12, 3538–3549.
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748.
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128.
- Tsui, C.; Kong, E.F.; Jabra-Rizk, M.A. Pathogenesis of Candida albicans biofilm. Pathog. Dis. 2016, 74, ftw018.
- Maza, P.K.; Bonfim-Melo, A.; Padovan, A.C.B.; Mortara, R.A.; Orikaza, C.M.; Ramos, L.M.D.; Moura, T.R.; Soriani, F.M.; Almeida, R.S.; Suzuki, E.; et al. Candida albicans: The ability to invade epithelial cells and survive under oxidative stress is unlinked to hyphal length. Front. Microbiol. 2017, 8.
- Galocha, M.; Pais, P.; Cavalheiro, M.; Pereira, D.; Viana, R.; Teixeira, M.C. Divergent approaches to virulence in C. Albicans and C. Glabrata: Two sides of the same coin. Int. J. Mol. Sci. 2019, 20, 2345.
- Lin, C.J.; Chen, Y.L. Conserved and divergent functions of the CAMP/PKA signaling pathway in candida albicans and candida tropicalis. J. Fungi 2018, 4, 68.
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68.
- Naglik, J.R.; Gaffen, S.L.; Hube, B. Candidalysin: Discovery and function in Candida albicans infections. Curr. Opin. Microbiol. 2019, 52, 100–109.
- Kasper, L.; König, A.; Koenig, P.A.; Gresnigt, M.S.; Westman, J.; Drummond, R.A.; Lionakis, M.S.; Groß, O.; Ruland, J.; Naglik, J.R.; et al. The fungal peptide toxin Candidalysin activates the NLRP3 inflammasome and causes cytolysis in mononuclear phagocytes. Nat. Commun. 2018, 9, 1–20.
- König, A.; Hube, B.; Kasper, L. The Dual Function of the Fungal Toxin Candidalysin during Candida albicans—Macrophage Interaction and Virulence. Toxins 2020, 12, 469.
- Priya, A.; Pandian, S.K. Piperine Impedes Biofilm Formation and Hyphal Morphogenesis of Candida albicans. Front. Microbiol. 2020, 11.
- Talapko, J.; Škrlec, I. The Principles, Mechanisms, and Benefits of Unconventional Agents in the Treatment of Biofilm Infection. Pharmaceuticals 2020, 13, 299.
- McCall, A.D.; Pathirana, R.U.; Prabhakar, A.; Cullen, P.J.; Edgerton, M. Candida albicans biofilm development is governed by cooperative attachment and adhesion maintenance proteins. NPJ Biofilms Microbiomes 2019, 5, 1–12.
- Cavalheiro, M.; Teixeira, M.C. Candida Biofilms: Threats, challenges, and promising strategies. Front. Med. 2018, 5, 28.
- Li, W.; Wang, J.J.; Qian, H.; Tan, L.; Zhang, Z.; Liu, H.; Pan, Y.; Zhao, Y. Insights into the Role of Extracellular DNA and Extracellular Proteins in Biofilm Formation of Vibrio parahaemolyticus. Front. Microbiol. 2020, 11.
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681.
- Nett, J.E.; Andes, D.R. Contributions of the biofilm matrix to candida pathogenesis. J. Fungi 2020, 6, 21.
- Paluch, E.; Rewak-Soroczyńska, J.; Jędrusik, I.; Mazurkiewicz, E.; Jermakow, K. Prevention of biofilm formation by quorum quenching. Appl. Microbiol. Biotechnol. 2020, 104, 1871–1881.
- Perry, A.M.; Hernday, A.D.; Nobile, C.J. Unraveling How Candida albicans Forms Sexual Biofilms. J. Fungi 2020, 6, 14.
- Glazier, V.E.; Murante, T.; Murante, D.; Koselny, K.; Liu, Y.; Kim, D.; Koo, H.; Krysan, D.J. Genetic analysis of the Candida albicans biofilm transcription factor network using simple and complex haploinsufficiency. PLoS Genet. 2017, 13, e1006948.
- Rodríguez-Cerdeira, C.; Martínez-Herrera, E.; Carnero-Gregorio, M.; López-Barcenas, A.; Fabbrocini, G.; Fida, M.; El-Samahy, M.; González-Cespón, J.L. Pathogenesis and Clinical Relevance of Candida Biofilms in Vulvovaginal Candidiasis. Front. Microbiol. 2020, 11, 2884.
- Bartnicka, D.; Karkowska-Kuleta, J.; Zawrotniak, M.; Satała, D.; Michalik, K.; Zielinska, G.; Bochenska, O.; Kozik, A.; Ciaston, I.; Koziel, J.; et al. Adhesive protein-mediated cross-talk between Candida albicans and Porphyromonas gingivalis in dual species biofilm protects the anaerobic bacterium in unfavorable oxic environment. Sci. Rep. 2019, 9.
- Ganguly, S.; Mitchell, A.P. Mucosal biofilms of Candida albicans. Curr. Opin. Microbiol. 2011, 14, 380–385.
- Murillo, L.A.; Newport, G.; Lan, C.Y.; Habelitz, S.; Dungan, J.; Agabian, N.M. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot. Cell 2005, 4, 1562–1573.
- Bonhomme, J.; d’Enfert, C. Candida albicans biofilms: Building a heterogeneous, drug-tolerant environment. Curr. Opin. Microbiol. 2013, 16, 398–403.
- Mallick, E.M.; Bergeron, A.C.; Jones, S.K.; Newman, Z.R.; Brothers, K.M.; Creton, R.; Wheeler, R.T.; Bennett, R.J. Phenotypic Plasticity Regulates Candida albicans Interactions and Virulence in the Vertebrate Host. Front. Microbiol. 2016, 7, 780.
- Brimacombe, C.A.; Sierocinski, T.; Dahabieh, M.S. A white-to-opaque-like phenotypic switch in the yeast Torulaspora microellipsoides. Commun. Biol. 2020, 3, 1–11.
- Wibawa, T. The role of virulence factors in Candida albicans pathogenicity. J. Med. Sci. 2016, 48, 58–68.
