Quorum sensing is a type of chemical communication by which bacterial populations control expression of their genes in a coordinated manner. This regulatory mechanism is commonly used by pathogens to control the expression of genes encoding virulence factors and that of genes involved in the bacterial adaptation to variations in environmental conditions. In phytopathogenic bacteria, several mechanisms of quorum sensing have been characterized. In this review, we describe the different quorum sensing systems present in phytopathogenic bacteria, such as those using the signal molecules named N-acyl-homoserine lactone (AHL), diffusible signal factor (DSF), and the unknown signal molecule of the virulence factor modulating (VFM) system. We focus on studies performed on phytopathogenic bacteria of major importance, including Pseudomonas, Ralstonia, Agrobacterium, Xanthomonas, Erwinia, Xylella,Dickeya, and Pectobacterium spp. For each system, we present the mechanism of regulation, the functions targeted by the quorum sensing system, and the mechanisms by which quorum sensing is regulated.
- : N-acyl-homoserine lactone
- diffusible signal factor
1. Introduction
Quorum sensing (QS) is a cell-to-cell communication mechanism used by bacteria for promoting collective behavior within a population. This cooperative behavior relies on the production, detection, and response to signal molecules in a cell-density-dependent manner. At a low cell density, a basal level of the signal molecule is produced by bacteria. Signal molecules can be diffused or exported into the extracellular environment. As bacterial density increases, signal molecules accumulate. After reaching a threshold, signal molecules are perceived by the bacteria, which initiate a set of biological activities in a coordinated fashion. Acyl-homoserine lactone (AHL) was the first signal molecule, identified in the 1980s[1][2]. Originally discovered in the bioluminescent marine bacterium Aliivibrio fischneri, these signal molecules were later characterized in a plethora of bacteria including Pectobacterium carotovorum (formerly named Erwinia), Agrobacterium tumefaciens, Citrobacter amalonaticus, and Pseudomonas aeruginosa[3][4]. Since then, several other types of QS signals have been identified, and most QS signals are either small organic molecules or peptides with 5 to 20 amino acids. In Gram-positive bacteria, the signal molecules are mainly peptides[5], while in Gram-negative bacteria, they are organic molecules smaller than 1000 Da. A universal signal described as autoinducer 2 (AI-2) is also produced by some Gram-positive and Gram-negative bacteria. These signal molecules are produced (i) at a specific growth stage, (ii) under particular physiological conditions, or (iii) in response to an environmental change. QS controls the expression of the many genes involved in a variety of functions, such as biofilm formation, toxin production, exopolysaccharide synthesis, extracellular enzyme production, motility, and plasmid conjugation. In pathogenic bacteria and, therefore, in plant pathogenic bacteria, QS plays a major role in the regulation of virulence factor productions and the infectious processes.
This entry aims to describe how phytopathogenic bacteria incorporate QS mechanisms into the complex regulatory cascades that control genes in pathogenicity and colonization of the host, and thereby update the data reviewed more than 15 years ago in Von Bodman et al. [6]. We present QS systems harbored by phytopathogenic bacteria, i.e., the ones relying on AHL or diffusible signal factors (DSF), in addition to the virulence factor modulating (VFM) system. For each of these systems, we present the regulatory mechanism, the target genes of QS, and the mechanisms that are involved in the QS process. Mansfield et al. previously listed 10 species of phytopathogenic bacteria of major economic and scientific importance[7]. Here, we focus on the QS systems present in these species, including Pseudomonas syringae, Ralstonia solanacearum, Agrobacterium tumefaciens spp., bacteria of the genus Xanthomonas spp., Erwinia amylovora, Xylella fastidiosa, Dickeya spp., and Pectobacterium spp. (Table 1).
Table 1. Quorum screening (QS) systems present in bacterial plant pathogen species. The table presents a ranked list of the bacteria according to Mansfield et al.
|
Top 10 Rank |
Bacterial Pathogen Species |
QS Mechanisms |
Involvement in Virulence |
|---|---|---|---|
|
1 |
Pseudomonas syringae |
AHL |
Yes |
|
2 |
Ralstonia solanacearum |
AHL |
No |
|
DSF-derived signals |
Yes |
||
|
3 |
Agrobacterium tumefaciens with pTi |
AHL |
Yes |
|
4 |
Xanthomonas oryzae pv oryzae |
DSF |
Yes |
|
5 |
Xanthomonas campestris pv |
DSF |
Yes |
|
6 |
Xanthomonas axonopodis pv |
DSF |
Yes |
|
7 |
Erwinia amylovora |
AHL |
Yes * |
|
8 |
Xylella fastidiosa |
DSF |
Yes |
|
9 |
Dickeya spp. |
AHL |
No ** |
|
Vfm |
Yes |
||
|
10 |
Pectobacterium carotovorum (and atrosepticum) |
AHL |
Yes |
* probably strain-dependent; ** species-dependent.
2. Diffusible Signal Factor-Mediated Quorum Sensing
Diffusible-Signal-Factor-Mediated QS is only present in three of the top 10 plant pathogenic bacteria: Xanthomonas oryzae pv. oryzae (Xoo), Xanthomonas campestriss (Xcc), Xanthomonas axonapodis, and Xylella fastidiosa. In addition, R. solanacearum can produce a DSF-derived signal molecule (Table 1).
2.1. Overview of DSF-Mediated Quorum Sensing
The Diffusible Signal Factor (DSF) family of signals features intriguing types of QS signal molecules found in diverse Gram-negative bacteria. Signal molecules are cis-2-unsaturated fatty acids that share a fatty acid carbon chain with variations in length, double-bond configurations, and side-chains[7]. Structural variants were mostly characterized using purification from culture supernatants followed by high performance liquid chromatography (HPLC) analyses and nuclear magnetic resonance (NMR). A much greater diversity of signals than previously anticipated was identified, including cis-2-dodecenoic acid (BDSF), cis, cis-11-methyldodeca-2,5-dienoic acid (CDSF), cis-2- and trans-2-decenoic acid (SDSF), cis-10-methyl-2-dodecenoic acid (IDSF or DSF-II), cis-9-methyl-2-decenoic acid, cis-2-undecenoic acid, 2-cis-unsaturated fatty acids (with the unsaturated fatty acids being 2-tetradecenoic acid (XfDSF1) or 2-cis-hexadecanoic acid (XfDSF2)), and 13-methyltetradecanoic acid (LeDSF3) (Figure 3, Table 3)[8]. A given organism can produce several signal molecules. Moreover, the growth environment affects the nature of the DSF variants[9][10].
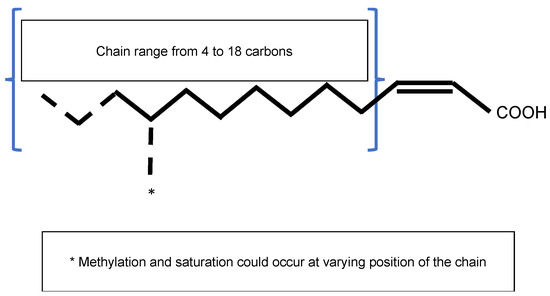
Figure 1. General structure of the Diffusible Signal Factor (DSF). Signal molecules are cis-2-unsaturated fatty acids. Fatty acid carbon chains vary in their lengths, double-bond configurations, and side-chain modifications, particularly methylation. Fatty acid carbon chains range from 8 to 14 carbons. A given species is able to produce different molecules. Methylation occurs at the first carbon for R. solanacearum signal molecules 3-OH-PAME or 3-OH-MAME.
Table 1. Overview of DSF-mediated quorum-sensing processes in phytopathogenic bacteria.
| Signal Molecule | Species | Studied Strains | QS System | Pathology | Hosts | Targeted Functions | References |
|---|---|---|---|---|---|---|---|
| OHHL OOHL |
Pseudomonas syringae pv. tabaci | 11528 | PsyI/PsyR | Wild-fire disease | Tobacco plants | Swarming, flagellum synthesis, assembly of pili, biofilm formation, chemotaxis, colonization, epiphytic viability, SST2, SST6, alginate synthesis | [11][12][13][14] |
| No production of AHL | Pseudomonas syringae pv. actinidiae | PsaR1, PsaR2, PsaR3 | Bacterial canker | Kiwifruit plants | Regulation of traits associated with survival in planta, cellular multiplication, swarming, oxidative stress resistance | [15][16] | |
| HHL C8-HSL |
Ralstonia solanacearum | GMI1000 | SolI/ SolR |
Wilting | No Data | [6][17][18][19] | |
| OOHL | Agrobacterium fabrum (tumefaciens) | C58 | TraI/ TraR |
Crown Gall | DNA replication, plasmid segregation in daughter cells, conjugative transfer of plasmid Ti | [20][21][22][23][24][25] | |
| OHHL HHL |
Erwinia amylovora | Ea2 | EamR/EamI | fire blight | Apple, Pear | Amylovoran, levan, tolerance to hydrogen peroxide | [26][27][28][29][30] |
| OHHL HHL DHL |
Dickeya dadantii | 3937 | ExpR/ExpI | Soft rot | Pineapple, Potato, Sweet potato, Banana, Maize, Dianthus spp., Philodendron, Pelargonium, Saintpaulia |
No implication in virulence | [31][32][33] |
| OHHL OOHL |
Dickeya zeae | EC1 | ExpR/ExpI | Soft rot | Maize, Potato, Pineapple, Banana, Tobacco, Rice, Brachiaria, Chrysanthemum | Swarming, pigment synthesis, cellular aggregate formation, plant colonization, rice seed germination No implication in PCWDE production | [34] |
| OHHL HHL |
Dickeya solani | ExpR/ExpI | Soft rot | Potato, Hyacinth | PCWDEs | [35] | |
| OHHL OOHL |
Pectobacterium carotovorum | ExpI/ExpR1—ExpR2 CarR/CarI | Soft rot | Potato, carrot, green pepper | PCWDEs, oxidative stress resistance, antimicrobial activity, carbapenem biosynthesis |
[4][6][36][37] | |
| OOHL C8-HSL OHHL |
Pectobacterium atrosepticum | ExpR/ExpI | Soft rot | Potato and chicory | Pectates lyases | [36][37][38][39][40][41] | |
| OHHL | Pantoea stewartiisubsp. stewartii | Leaf blight and Stewart disease | Corn | Mobility, stewartan production, carotinoids pigments | [42][43] |
OHHL: N-(3-oxohexanoyl)-HSL; HHL: N-(hexanoyl)-HSL; DHL: N-(decanoyl)-HSL; OOHL: N-(3-oxo-octanoyl)-HSL; HSL: Homoserine Lactone; SST2 and SST6: secretion system type 2 and 6, respectively; PCWDE: plant cell wall degrading enzymes.
Three different types of DSF-mediated QS systems were defined. Classification depends on the genomic context of the involved genes. While the first group contains DSF systems whose genes encoding key signaling components are colocalized on the genome, systems belonging to the second group gather genes that are not clustered in the genome. Finally, the third class contains DSF systems whose genes are not clearly identified[8]. Systems belonging to the first group were first identified and characterized in the phytopathogen Xanthomonas campestris pv. campestris (Xcc), which is responsible for black rot in crucifers. To date, every DSF system identified in plant pathogenic bacteria belongs to this first class. These DSF systems have also been studied in other Xanthomonas species and in Xylella fastidiosa.
Briefly, three genes named rpfF, rpfC, and rpfG encode the main components of the DSF biosynthetic pathway, which depends on fatty acid biosynthesis. RpfF is a DSF synthase, and RpfC–RpfG is a two-component regulatory system involved in signal perception and transduction. RpfF is a bifunctional enzyme with thioesterase activity that first cleaves the thioester bonds of acyl-ACPs to release holo–ACPs, and then its enoyl-CoA hydratase activity dehydrates the holo–ACP substrates to the final product[10] RpfF is active towards acyl-ACP substrates, with carbon chains ranging from 8 to 14. A given RpfF protein is able to produce multiple DSF signals[44][45]. RpfC is the DSF sensor, composed of a transmembrane domain (TM), an histidine kinase domain (HK), a receiver domain (REC), and a histidine phosphotransferase domain (HTP)[7]. The mechanism by which DSF is detected by this sensor is still unknown, but the sensor uses a phospho-relay mechanism to transfer the signal to the response regulator RpfG (Figure 2) [8][46][47][48]. The RpfG N-terminal response regulator (RR) domain interacts directly with RpfC, whereas its HD-GYP domain has phosphodiesterase activity that is activated by the DSF signal. This domain degrades cyclic di-GMP into two GMP molecules. Cyclic di-GMP binds to the global transcription factor Clp and represses rpfB expression. When cyclic di-GMP is degraded, free forms of Clp dominate, which drives the expression of several hundred genes, including those encoding virulence factors[49].
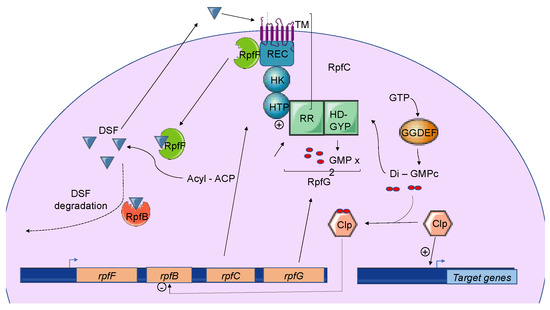
Figure 2. The Diffusible Signal Factor-mediated quorum-sensing (DSF-QS) system. In phytopathogenic bacteria, the DSF system is encoded by the rpf gene cluster. RpfF is a bifunctional enzyme involved in the production of DSF molecules. RpfB is proposed to be involved in DSF turnover. RpfC–RpfG is a two-component regulatory system that is involved in signal perception and transduction. RpfC is a DSF sensor that uses a phospho-relay mechanism to transfer the signal to the response regulator, RpfG. The N-terminal RR response domain of RpfG interacts directly with RpfC. Its HD-GYP domain then degrades cyclic di-GMP. RpfC can also bind to RpfF using its C-terminal REC domain and negatively regulates DSF biosynthesis. At a low cell density, (i) RpfC forms a complex with RpfF, blocking its enzymatic activity and inhibiting DSF signal biosynthesis, and (ii) cyclic di-GMP binds to the global transcription factor Clp, which represses rpfB expression. At a high cell density, RpfF is released and produces DSF signals, which allow the induction of QS regulation. Cyclic di-GMP is degraded by the HD-GYP domain of RpfG, and rpfB is expressed, like several genes encoding virulence factors activated by Clp.
2.2. Functions Regulated by DSF-Mediated Quorum Sensing in Plant Pathogens
2.2.1. Xanthomonas spp.
Including X. oryzae, X. campestris, and X. axonopodisXanthomonas spp. are yellow-pigmented bacteria, several species of which are phytopathogens associated with economically important crops worldwide. Bacteria from this genus are responsible for diseases on approximatively 400 plant species (124 monocots and 268 dicots), including rice, wheat, citrus, tomato, pepper, banana, and bean. There is a high degree of specificity between the host plant and the Xanthomonas species and pathovars[50]. Infection is divided into two stages: the epiphytic stage and the endophytic stage. Briefly, the pathogen penetrates into a new host via plant natural openings and wounds, colonizes the host, and invades either the vascular system or the intercellular spaces of the mesophyll parenchyma tissue. Virulence factors are mainly type III secretion systems (T3SS) and the effectors secreted by these systems. In addition, other pathogenicity factors have been involved, directly or indirectly, in the virulence, such as PCWDEs, type IV secreted effectors, adhesins, and lipopolysaccharides[51]. It has been clearly demonstrated that Xcc recruits DSF to synchronize virulence gene expression. Transcriptome analyses of mutant strains modified in the production of DSF led to the identification of 165 genes whose expression is significantly varied. Among these genes are those involved in extracellular enzyme and extracellular polysaccharide production, flagella synthesis, iron uptake, aerobic respiration, resistance to toxins, and oxidative stress[52]. Phenotypes associated with these genes are complemented by the addition of DSF signals[52]. Additionally, DSF is involved in biofilm formation[47]. DSF has also been implicated in the intricate crosstalk between Xanthomonas spp. and their host plants. DSF facilitates Xanthomonas spp. entry into host plants [53]. DSF elicits innate immunity and the secretion of xanthan, the main exopolysaccharide that suppresses the DSF-induced innate immunity in Xcc[54]. Taken together, these indicate that plants have evolved to recognize a widely conserved bacterial communication system [54].Finally, studies of the DSF system in Xoo, the causative agent of rice bacterial blight disease, and in Xanthomonas axonopodis pv. glycines (Xag), which is responsible for the bacterial pustule of soybean, demonstrated that the mechanisms for DSF biosynthesis and regulation are conserved and promote the regulation of virulence factor productions or factors associated with virulence in both pathovars[55][56]. In Xag, the regulated extracellular enzymes are mainly carboxyl–methyl cellulases, proteases, endo-β-1,4-mannanases, and pectate lyases[56].
2.2.2. In Xylella fastidiosa
Xyllella fastidiosa, which is divided into four subspecies, belongs to the Xanthomonadaceae genus[57]. Xyllella fastidiosa is a vascular wilt pathogen that has a wide host range of over 350 plant species, including almond leaves, lemons, and olive trees. X. fastidiosa is always found in the xylem tissue of its plant host or in the foregut of its xylem-feeding hemipteran insect vector. In contrast to other Xanthomonads, X. fastidiosa does not carry T3SS, and the symptoms associated with this pathogen depend on the capacity of the bacteria to colonize and block xylem vessels, leading to drought stress. Hence, attachment, motility, and biofilm formation are important for infection[57]. A study of rpfF and rpfC mutants in X. fastidiosa showed that switching between the plant host and insect vectors is regulated by QS (Table 3). DSF-mediated QS also controls virulence factors, such as adhesins, migration in xylem vessels, and the colonization of insect vectors [56][58]. At a high cell density, DSF-mediated QS promotes cell stickiness, thereby encouraging attachment to the xylem wall[56]. In addition, outer membrane vesicle (OMV) production is repressed via DSF by an unknown mechanism[59]. OMVs block bacterial interactions with the xylem vessel walls. Taken together, this interaction indicates that DSF-mediated QS favors X. fastidiosa attachment to the vessels. This phenomenon has been proposed to help bacterial acquisition by insects[58]].In summary, X. fastidiosa coordinates its behavior according to its infection stages (insect vectors vs. plant host) and population size using a DSF-mediated QS similar to that present in Xanthomonas spp. [58].
2.2.2. Other DSF-Derived Signals
As mentioned before, the R. solanacearum species complex carries two QS systems—one depending on AHL signals, which does not seem to be involved in the regulation of virulence, and a second one whose signal molecules are close to the DSF family. This system regulates the virulence of R. solanacearum species [60]. The R. solanacearum species complex is divided into two clades, depending on their QS signal types, with one being the (R)-methyl 3-hydroxymyristate (or 3-OH-MAME) and the second one being (R)-methyl 3-hydroxypalmitate (or 3-OH-PAME)[61]. PhcB methyltransferases synthesize both QS signals from the cognate fatty acids, but the specific production of signals depends on the strains [62]. The QS signal is recognized by the two-component regulatory system PhcS–PhcR. In the absence of a signal, PhcR inhibits the PhcA regulator, which activates the production of exopolysaccharides and PCWDEs. In the presence of the signal, PhcR is phosphorylated by PhcS, which inhibits it, and thereby increases the number of functional PhcA and the production of virulence factors[60]. In this way, early and late virulence factors are coordinately controlled by cell density, allowing the proper expression of genes encoding virulence factors, which is important for the success of the infectious process.
2.2. Induction, Maintenance, and Turnover of DSF-Mediated Quorum Sensing
2.2.1. Induction and Maintenance
In Xcc, DSF biosynthesis is autoregulated by a posttranslational mechanism. In addition to its interaction with RpfG, RpfC can also bind to RpfF using its C-terminal REC domain and negatively regulate DSF biosynthesis[7][8] (Figure 2). At low cell density, unphosphorylated RpfC conformation facilitates the formation of the RpfC–RpfF complex blocking the enzymatic activity of RpfF and the production of DSF signals. At a higher cell density, RpfF is released and produces DSF signals, which allows the induction of QS regulation [51]. This mechanism demonstrated in Xcc seems also to be present in X. fastidiosa.
| Top 10 Rank [7] | Bacterial Pathogen Species | QS Mechanisms | Involvement in Virulence |
|---|---|---|---|
| 1 | Pseudomonas syringae | AHL | Yes |
| 2 | Ralstonia solanacearum | AHL | No |
| DSF-derived signals | Yes | ||
| 3 | Agrobacterium tumefaciens with pTi | AHL | Yes |
| 4 | Xanthomonas oryzae pv oryzae | DSF | Yes |
| 5 | Xanthomonas campestris pv | DSF | Yes |
| 6 | Xanthomonas axonopodis pv | DSF | Yes |
| 7 | Erwinia amylovora | AHL | Yes * |
| 8 | Xylella fastidiosa | DSF | Yes |
| 9 | Dickeya spp. | AHL | No ** |
| Vfm | Yes | ||
| 10 | Pectobacterium carotovorum (and atrosepticum) | AHL | Yes |
2. Diffusible Signal Factor-Mediated Quorum Sensing
2.1. Overview of DSF-Mediated Quorum Sensing


2.2. Functions Regulated by DSF-Mediated Quorum Sensing in Plant Pathogens
2.2.1. Xanthomonas spp. Including X. oryzae, X. campestris, and X. axonopodis
2.2.2. In Xylella fastidiosa
2.2.2. Other DSF-Derived Signals
2.2. Induction, Maintenance, and Turnover of DSF-Mediated Quorum Sensing
2.2.1. Induction and Maintenance
2.2.2. Turnover of DSF Signals
In Xcc and Xoo, DSF signals accumulate in the early stationary phase, and their levels decline rapidly afterwards, suggesting the existence of a DSF signal turnover system[61]. Studies of RpfB in both Xcc and X. fastidiosa have shown that RpfB is involved in DSF processing, as DSF-like fatty acid profiles whose production depends on RpfF are affected in rpfB mutants[9]. In addition, rpfB mutants boost DSF production during growth, while the overproduction of RpfB abolishes the DSF signal[49]. A reduction in insect colonization and transmission was observed, but not a reduction in plant colonization. A biochemical analysis performed in vitro suggested fatty acyl-CoA ligase activity for RpfB, but, surprisingly, its effects on the DSF and BDSF signals was limited, indicating that RpfB plays a more important role in pathogenesis by counteracting RpfF thioesterase activity [63] Discrepancies in RpfB enzymatic activities measured in vitro and in vivo suggest the involvement of an additional factor.
The expression of rpfB is negatively regulated by RpfC, RpfG, and Clp, which directly bind to the rpfB promoter region when it is complexed with di-GMP-cyclic [64] (Figure 2). At a low cell density, the di-GMP-cyclic-Clp complex represses rpfB expression, whereas at a high cell density, di-GMP-cyclic is degraded by RpfG, and rpfB is expressed. Finally, the RpfB-dependent signal turnover system was also detected in several Xanthomonas spp. including Xoo, but discrepancies were observed in bacterial virulence-associated traits[49].
