MicroRNA (miRNAs) are short, single-stranded ncRNAs that can be measured in biofluids as minimally invasive biomarkers of disease, including cancer. Their reliable measurement however is challenging, therefore several methods are being developed for miRNA quantitation. Here the technologies currently in use or being developed for quantitation of miRNAs are described.
- MicroRNA
- biofluids
- methods
- biomarkers
- cancer
1. Introduction
from: Precazzini F, Detassis S, Imperatori AS, Denti MA, Campomenosi P. Measurements Methods for the Development of MicroRNA-Based Tests for Cancer Diagnosis. Int J Mol Sci. 2021 Jan 25;22(3):1176.
Blood circulating microRNAs (c-miRNAs) are considered valuable biomarkers since they have show high stability, resistance to ribonucleases and to severe physicochemical conditions in body fluids. This favours their use in clinical applications. Differential expression of c-miRNAs showed big potential for cancer screening. Thus, these small RNA molecules may function as clinical biomarkers to identify the presence of tumors, to select treatment strategies and to predict outcomes as explained in Figure 1. Composition and levels of extracellular miRNAs can vary as a consequence of diseases or other conditions [1]. Several reports show that miRNAs are sensitive to physiological or pathological changes, specific to the pathology of interest, and they represent a reliable indicator of the presence of disease before clinical symptoms appear [2]. Moreover miRNAs have shown great potential as diagnostic biomarkers for neurodegenerative disorders, cardiovascular diseases, endometriosis, cancer and several other diseases.
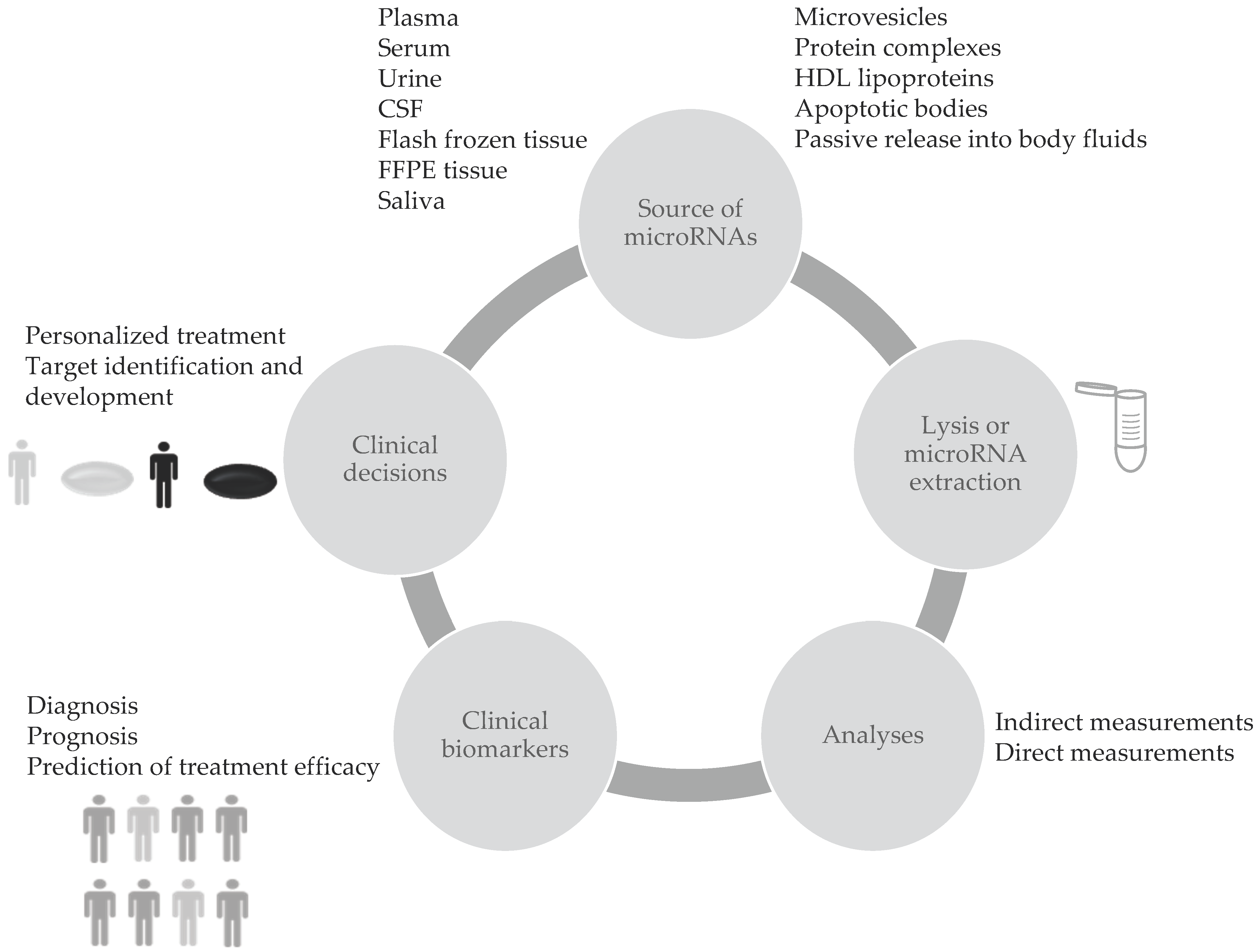
2. Technical Llimitations in Mmeasurement of c-miRNAs as Bbiomarkers
If biological complexity is already difficult to capture, there are also technical limitations in c-miRNAs measurement that make it difficult to develop a diagnostic or prognostic test directly translatable in clinical settings. In fact, despite the growing interest of the scientific community, proved by the enormous amount of articles published in recent years (a search for “circulating microRNAs AND cancer biomarker” on PubMed, performed in September 2020, yielded 123 entries in 2012, 189 in 2013, 254 in 2014, 335 in 2015, 329 in 2016, 338 in 2017, 356 in 2018 and 310 in 2019) only two miRNA based diagnostic tests (Biomild, Cosmos II) have reached phase 4 in clinical trials [3]. The main reasons for the limited clinical application may be classified into three categories: 1) pre-analytical, 2) analytical and 3) post-analytical factors. These factors affect measurement accuracy and reproducibility independently of its clinical application. .
If biological complexity is already difficult to capture, there are also technical limitations in c-miRNAs measurement that make it difficult to develop a diagnostic or prognostic test directly translatable in clinical settings. In fact, despite the growing interest of the scientific community, proved by the enormous amount of articles published in recent years (a search for “circulating microRNAs AND cancer biomarker” on PubMed, performed in September 2020, yielded 123 entries in 2012, 189 in 2013, 254 in 2014, 335 in 2015, 329 in 2016, 338 in 2017, 356 in 2018 and 310 in 2019) only two miRNA based diagnostic tests (Biomild, Cosmos II) have reached phase 4 in clinical trials. The main reasons for the limited clinical application may be classified into three categories: 1) pre-analytical, 2) analytical and 3) post-analytical factors. These factors affect measurement accuracy and reproducibility independently of its clinical application. .
.
| Phase of the Analysis | Challenges |
|---|---|
| Pre-analytical phase |
|
| Analytical phase |
|
| Post-analytical phase (normalization) |
|
3. Indirect Mmethods for miRNA Mmeasurement, Rrelying on RNA Eextraction & Rreverse Ttranscription (RT)
Several methods have been applied to quantify c-miRNAs, including quantitative real-time PCR, digital PCR, microarray, and high-throughput small RNA-sequencing. These methods need RNA isolation, in some instances small RNA enrichment, and/or reverse transcription before proceeding to miRNA quantification. A summary of the different techniques described in this review is depicted in Figure 2.
Several methods have been applied to quantify c-miRNAs, including quantitative real-time PCR, digital PCR, microarray, and high-throughput small RNA-sequencing. These methods need RNA isolation, in some instances small RNA enrichment, and/or reverse transcription before proceeding to miRNA quantification. A summary of the different techniques described in this review is depicted in figure 2.
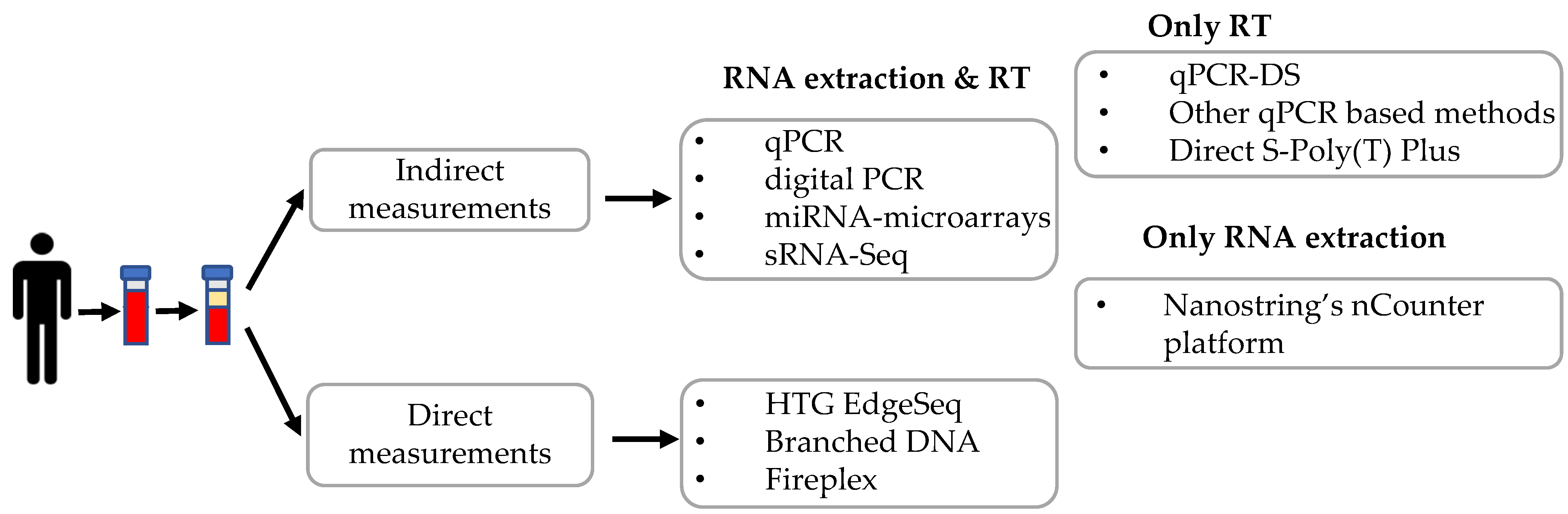
Graphical summary of the different techniques described in this review. RT: reverse transcription.
4. Direct Mmethods for miRNA Mmeasurement from Bbiofluids
HTG EdgeSeq
In 2018 an approach called HTG EdgeSeq, developed by HTG Molecular Diagnostic, Inc. (Tucson, AZ, USA), was described for the direct screening of plasma c-miRNAs. By eliminating RNA extraction and reverse transcription, it simplified both library and sample preparation for RNA and miRNA targeted sequencing. Probes are used to form double stranded molecules with targets and protect the complexes from degradation with S1 nuclease. The probes also carry “wings”, extensions that will allow introduction of adapters for NGS by PCR, protected by “wingmen”. Single stranded nucleic acids (targets and unbound probes), will be degraded, leaving the target-probe complexes available for subsequent steps (Figure 3). This approach allows direct quantification of transcripts or miRNAs, allowing to increase the throughput and to considerably reduce bioinformatic analyses and time to results (~36 hours). Profiles of more than 2000 miRNA transcripts can be performed starting from a relatively small amount of initial sample. Normalization can be performed as classical sRNA-Seq. Moreover, the HTG EdgeSeq costs are consistent with other next-generation sequencing (NGS) approaches. In a study by Songia and coworkers peripheral blood from four healthy subjects was collected. Samples were barcoded, purified and quantified using a KAPA Library Quantification kit [4]. Overall HTG-sequencing with Illumina NextSeq was performed leading to the evaluation of the expression of 2083 miRNAs starting from just 15 μL of plasma. In addition, the study compared the performances of techniques that are commonly used for miRNA detection like quantitative PCR (qPCR) and the chip-based digital PCR (dPCR) and a strong correlation between qPCR and dPCR with HTG was found [4]. In another comparative study, the performance of HTG Edgeseq was compared with that of classical RNA-seq, Fireplex and Nanostring nCounter platforms in measurement of synthetic miRNA pools or miRNAs from plasma samples [146]. HTG Edgeseq showed the smallest coefficient of variation when repeated measures were performed and low bias in measuring “ratiometric pools” of synthetic miRNAs. Moreover, it could detect variation in the levels of placenta-specific miRNAs between pregnant and non-pregnant women [5].
In 2018 an approach called HTG EdgeSeq, developed by HTG Molecular Diagnostic, Inc. (Tucson, AZ, USA), was described for the direct screening of plasma c-miRNAs. By eliminating RNA extraction and reverse transcription, it simplified both library and sample preparation for RNA and miRNA targeted sequencing. Probes are used to form double stranded molecules with targets and protect the complexes from degradation with S1 nuclease. The probes also carry “wings”, extensions that will allow introduction of adapters for NGS by PCR, protected by “wingmen”. Single stranded nucleic acids (targets and unbound probes), will be degraded, leaving the target-probe complexes available for subsequent steps (Figure 8). This approach allows direct quantification of transcripts or miRNAs, allowing to increase the throughput and to considerably reduce bioinformatic analyses and time to results (~36 hours). Profiles of more than 2000 miRNA transcripts can be performed starting from a relatively small amount of initial sample. Normalization can be performed as classical sRNA-Seq. Moreover, the HTG EdgeSeq costs are consistent with other next-generation sequencing (NGS) approaches. In a study by Songia and coworkers peripheral blood from four healthy subjects was collected. Samples were barcoded, purified and quantified using a KAPA Library Quantification kit. Overall HTG-sequencing with Illumina NextSeq was performed leading to the evaluation of the expression of 2083 miRNAs starting from just 15 μL of plasma. In addition, the study compared the performances of techniques that are commonly used for miRNA detection like quantitative PCR (qPCR) and the chip-based digital PCR (dPCR) and a strong correlation between qPCR and dPCR with HTG was found. In another comparative study, the performance of HTG Edgeseq was compared with that of classical RNA-seq, Fireplex and Nanostring nCounter platforms in measurement of synthetic miRNA pools or miRNAs from plasma samples. HTG Edgeseq showed the smallest coefficient of variation when repeated measures were performed and low bias in measuring “ratiometric pools” of synthetic miRNAs. Moreover, it could detect variation in the levels of placenta-specific miRNAs between pregnant and non-pregnant women.
Figure 3. Diagram of HTG Edgeseq workflow. The hybridization, S1 nuclease treatment and PCR steps are depicted. These steps allow it to protect probes bound to target miRNAs, removing other RNAs and unbound probes. Wash steps are not shown.
8. Diagram of HTG Edgeseq workflow. The hybridization, S1 nuclease treatment and PCR steps are depicted. These steps allow it to protect probes bound to target miRNAs, removing other RNAs and unbound probes. Wash steps are not shown.
Table 2. Summary of the different time, throughput and multiplexing of the different methods described in this entry.
| Method | Time | Throughput | Multiplexing | ||
|---|---|---|---|---|---|
| INDIRECT MEASUREMENTS | RNA extraction and RT | qPCR | <6 h | Medium | Low |
| digital PCR | <6 h | Medium | Low | ||
| Microarray | ~2 days | Low | High | ||
| NGS | 1–2 weeks | Medium | High | ||
| RT only | RT-qPCR-DS | <4 h | Medium | Low | |
| Cell to Ct RT-qPCR | |||||
| Direct S-Poly(T) Plus | <4 h | Medium | Low | ||
| RT-qPCR | <4 h | Medium | Low | ||
| RNA extraction only | Nanostring Platform | ~2 days | Low | High | |
| DIRECT MEASUREMENTS | HTG EdgeSeq | ~36 h | Medium | High | |
| bDNA | ~12 h | Low | High | ||
| Fireplex | ~6 h | High | Medium | ||
| Chem-NAT | ~3 h | Medium | Low |
5. Conclusions
A comprehensive knowledge of c-miRNAs and their origin, as well as of other classes of non-coding RNAs including lncRNAs and circular-RNAs, has still to be obtained. The combination of miRNAs with other biomarkers should be evaluated. However, a clinical diagnostic test requires not only specific analytes that can identify the presence of a condition, be it cancer or other diseases, but also a reliable method for their quantification. The present review describes up to date techniques that can be applied, and whose performance can be compared, to address the issue of reliable miRNA biomarker quantification. With stringent guidelines, a detailed description of the procedures used for miRNA quantification in published papers and increased application of direct measurement methods, we expect that a step closer to their clinical application can be reached. Independent validation studies are required before microRNA measurement can be translated into clinical applications.
A comprehensive knowledge of c-miRNAs and their origin, as well as of other classes of non-coding RNAs including lncRNAs and circular-RNAs, has still to be obtained. The combination of miRNAs with other biomarkers should be evaluated. However, a clinical diagnostic test requires not only specific analytes that can identify the presence of a condition, be it cancer or other diseases, but also a reliable method for their quantification. The present review describes up to date techniques that can be applied, and whose performance can be compared, to address the issue of reliable miRNA biomarker quantification. With stringent guidelines, a detailed description of the procedures used for miRNA quantification in published papers and increased application of direct measurement methods, we expect that a step closer to their clinical application can be reached. Independent validation studies are required before microRNA measurement can be translated into clinical applications.
References: plesa




