Lysosomes, the membrane-bound digestive cell organelles, are well-known for their catabolic function.
- endolysosome
- ion channel
1. Introduction
Lysosomes, the membrane-bound digestive cell organelles, are well-known for their catabolic function. They dynamically interact with autophagy and endocytosis pathways which transport intracellular components or extracellular cargos to lysosomes, respectively [1][2][3][4] [1,2,3,4] (Figure 1). Both intracellular and extracellular cargos can be broken down into their constituent building blocks (e.g., amino acids and free fatty acids) by lysosomes through hydrolysis [5,6][5][6]. To date, around 60 hydrolytic enzymes, including proteases, lipases and nucleases have been found in lysosomes [7]. The activity of these hydrolytic enzymes is stimulated by the acidic lumen (pH 4.5–5.5) of lysosomes, which is established by the vacuolar H+-ATPase (V-ATPase), an ATP-driven proton pump [5,8,9][5][8][9] (Figure 2). Metabolites produced by hydrolysis are eventually recycled back to the cytoplasm or exported to the extracellular environment through exocytosis (Figure 1).
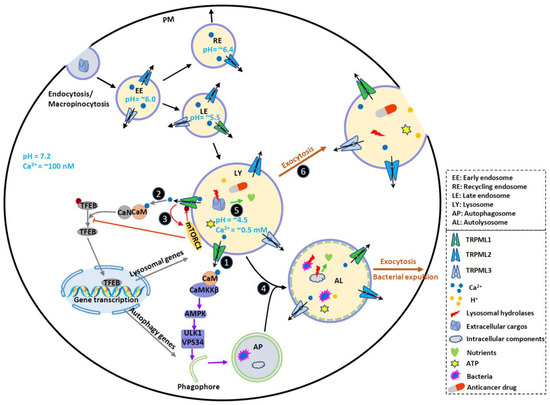
Figure 1. TRPMLs in endocytic, phagocytic and autophagic pathways and tumor progression. TRPMLs are predominately located on the endolysosomal pathway, and they have been involved in endocytic, phagocytic and autophagic pathways. In the tumor microenvironment, autophagy is activated to help cancer cells digest damaged or nonessential proteins and organelles, meeting the increased energy and nutrient demand of cancer cells. By releasing intraluminal Ca2+, TRPMLs have been implicated in intracellular Ca2+ signaling, endolysosome trafficking, and lysosomal functions, further regulating autophagy. First, TRPML1 activates calmodulin (CaM)/CaMKKβ/AMPK pathway to promote autophagosome formation. Second, TRPML1 activates CaM/CaN/TFEB pathway to continuously supply lysosome and autophagy proteins. Third, TRPML1 maintains mTORC1 activity to prevent cancer cell death and promote lysosome reformation. In normal fed conditions, mTORC1 phosphorylates TRPML1 at S571 and S576 to inhibit its activity. Starvation reduces mTORC1 activity and disinhibits TRPML1. This subsequently promotes mTORC1 activity, preventing cell death. Fourth, TRPML1 stimulates Apoptosis-linked gene-2 (ALG-2)-dependent lysosome centripetal movement to facilitate autophagosome-lysosome fusion. Fifth, TRPML1 increases lysosomal degradative functions, likely through controlling lysosomal pH. Sixth, TRPML1 increases Syt7-dependent lysosomal exocytosis, releasing hydrolases, ATP and H+ to extracellular spaces. These may change tumor microenvironment, promote ECM degradation, and facilitate tumor progression. By promoting lysosomal exocytosis, TRPML1 may also participate in drug resistance by releasing sequestrated anticancer drugs. Due to the functional redundancy between the TRPML proteins, TRPML2 and TRPML3 may also contribute to some of these events to regulate cancer development. Therefore, TRPMLs orchestrate all these cellular events to help cancer cells maintain high autophagic flux and adapt to the tumor microenvironment.
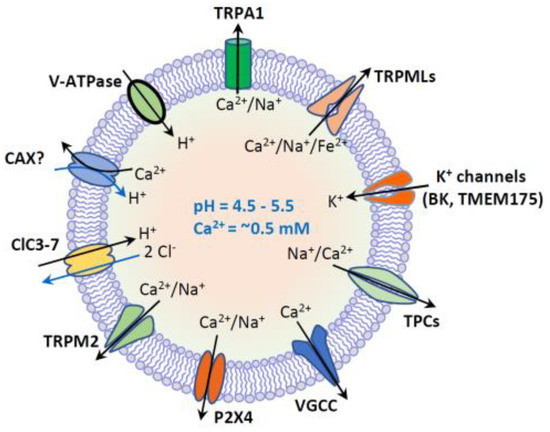
Figure 2. Lysosome ion homeostasis and ion channels. The lysosome has an acidic lumen that contains soluble hydrolytic enzymes. The activity of hydrolytic enzymes is controlled by intraluminal ion homeostasis that is established by multiple ion channels and transporters. These ion channels and transporters include H+-ATPase, nonselective cation channels (TRPML1-3, TRPM2, TRPA1 and P2 × 4), Na+ or Na+/Ca2+-selective two-pore channels (TPC1-3), voltage-gated Ca2+ channels (VGCC), K+-selective channels (BK and TMEM175), and 2Cl−/1H+ exchanger or Cl−channels (ClC3-7). Putative Ca2+/H+ exchanger (CAX) or Ca2+ transport protein mediates lysosomal uptake of Ca2+. Lysosomal Ca2+ (~0.5 mM) is important for membrane trafficking, and lysosomal pH (4.5–5.5) is essential for the activity of hydrolytic enzymes.
In addition to being the degradative endpoints, lysosomes are also the cellular signaling hubs that are involved in the regulation of cell growth, proliferation and differentiation [6,7,10,11][6][7][10][11]. For example, the lysosomal membrane hosts nutrient- and energy-sensing machineries in response to both internal stimuli and external changes of environment. In particular, both the master nutrient sensor mechanistic target of rapamycin complex 1 (mTORC1) [6][12][13][14][15][16] [6,12,13,14,15,16] and the master energy sensor AMP-activated protein kinase (AMPK) [17][18][19][20] [17,18,19,20] are associated with lysosomes and strictly control cell growth, proliferation and differentiation by sensing cellular metabolic status [17,21][17][21].
Lysosomal degradation and signaling require the establishment of the luminal ionic homeostasis [1] [1] including Ca2+, Na+, K+, Cl− and heavy trace metals such as Fe2+ and Zn2+. The lysosome is one of the main storage organelles of the second messenger Ca2+. The luminal Ca2+ concentration of lysosome is ~0.5 mM [22,23][22][23], which is approximately 5000-fold higher than that in cytosol (~100 nM) [24] [24] (Figure 1). The Ca2+ gradient across the lysosomal membrane is important, as it enables a small fraction of lysosomal Ca2+ efflux to generate a marked signal, resulting in the activation of downstream signaling cascades [2,25,26,27][2][25][26][27]. Lysosomes express various Ca2+-permeable channels on their membranes to regulate both local and global intracellular Ca2+ signals. These Ca2+-permeable channels include TRP Mucolipins (TRPMLs, TRPML1-3), Two Pore Channels (TPCs, TPC1-2), TRP Melastatin 2 (TRPM2), TRP Ankyrin 1 (TRPA1), P2X4 purinoceptor, and Voltage-Gated Ca2+ Channel (VGCC) [1,23,28,29,30,31,32][1][23][28][29][30][31][32] (Figure 2).
2. Endolysosomal TRPMLs
TRPMLs are the most intensively studied lysosomal Ca2+ channels, which belong to the large family of TRP ion channels [33][34][35] [33,34,35] (Figure 3). Compared with other channels of TRP family that are expressed on plasma membrane (PM), TRPMLs act predominantly in the endolysosomal system, regulating vesicles trafficking and function along endolysosomal pathways. All three members of TRPMLs can be activated by PI (3,5) P2, an endolysosome-specific phosphoinositide [36]. In mammals, TRPML1 is expressed in all tissues [37[37][38],38], and it is predominately localized on the late endosome (LE) and lysosome [35,39][35][39] where the low luminal pH facilitates its activation [39]. By releasing lysosomal Ca2+, TRPML1 regulates several membrane-trafficking processes, including lysosome to trans-Golgi–network (TGN) retrograde trafficking, autophagosome-lysosome fusion, and lysosomal exocytosis [1,40,41,42][1][40][41][42]. Mutations in TRPML1 causes a human autosomal recessive disease named mucolipidosis type IV (MLIV), a lysosomal storage disease (LSD) [37,43][37][43]. Impaired TRPML1 has also been associated with several other LSDs [44]. Compared with TRPML1, TRPML2 and TRPML3 are less understood. Both of them are more easily activated by higher pH [45,46,47][45][46][47]. Although TRPML2 is found in most organs, it is abundant in immune cells and tissues [48,49,50][48][49][50]. Subcellularly, TRPML2 is primarily expressed on the recycling endosomes (RE) and the early endosomes (EE) [46][50][51] [46,50,51] where it regulates the recycling of specific proteins from RE/EE to the cell surface [51]. Emerging evidence suggests that TRPML2 is an osmo/mechanosensitive ion channel on endolysosomal membranes [52]. Currently, TRPML2 has not been linked to any human disorders. However, it may play an important role in the secretion of chemokine and cytokine by macrophages [46,50][46][50]. TRPML3 is predominantly localized in the endocytic and autophagic pathways, in line with its cellular function of regulating endocytosis and autophagy [53,54,55][53][54][55]. The expression of TRPML3 has been detected in skin melanocytes, hair cells of the inner ear, neonatal intestinal enterocytes, as well as cells in the thymus, kidney and lung [56,57][56][57]. The gain-of-function mutation A419P in TRPML3 causes the varitint-waddler phenotype in mice, characterized by hearing loss and circling behavior [47,58,59][47][58][59].
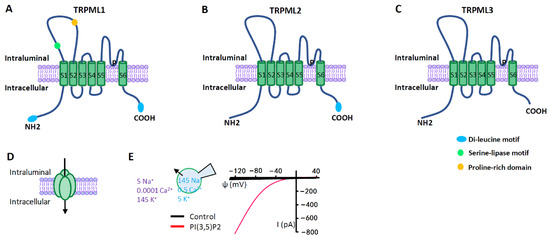
Figure 3. TRPML ion channels. (A–C) TRPML pore-forming subunit contains six transmembrane segments (S1–S6) and a putative pore region (P), with presumably cytosolic N- and C- termini. Distinct from other TRP channels, TRPMLs are characterized by a large extracellular (or intraluminal) loop between S1 and S2. There are dileucine motifs in TRPML1 and TRPML2 at their C- and/or N- termini to determine their intracellular endolysosomal localization. The endolysosomal localization of TRPML3 is determined by its heteromultimerization with other TRPMLs. (D) Functional TRPMLs are tetramers. (E) TRPMLs currents measured using lysosome-patch-clamp method. Physiological asymmetric solutions are used. The bath solution (cytosolic) contains (in mM) 145 K+, 5 Na+, 0.0001 Ca2+, and pH 7.2. Pipette solution (luminal) contains (in mM) 145 Na+, 5 K+, 0.5 Ca2+, and pH 4.6. TRPMLs are activated by cytosolic PI (3,5) P2 (1 µM), a lysosome-specific phosphoinositide. TRPMLs are inwardly (cation flowing from the lumen to the cytosol) rectifying channels permeable to cations. Given the topology of the TRPML proteins at the lysosomal membrane and the electrical properties of the lysosome, TRPML opening leads to Ca2+ and Na+ release from the lysosome to the cytosol.
Given the crucial role of lysosomes in multiple biological functions and signals, dysregulation of lysosomal function may cause human diseases. Indeed, a growing number of studies have demonstrated that tumor angiogenesis and progression are associated with altered lysosomes [60,61,62,63][60][61][62][63]. Growing evidence also suggests that intracellular Ca2+ signals mediated by lysosomal Ca2+ channels may control the development of various cancers. Herein, we summarize the emerging role of lysosomes and TRPMLs in cancer development. We hope to guide the readers into a more in-depth discussion of the relationship between lysosomal Ca2+ signaling and cancer development, potentially directing the development of new therapeutics for cancer.
References
- Xu, H.; Ren, D. Lysosomal Physiology. Annu. Rev. Physiol. 2015, 77, 57–80, doi:10.1146/annurev-physiol-021014-071649.
- Luzio, J.P.; Pryor, P.R.; Bright, N.A. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 2007, 8, 622–632, doi:10.1038/nrm2217.
- Kolter, T.; Sandhoff, K. PRINCIPLES OF LYSOSOMAL MEMBRANE DIGESTION: Stimulation of Sphingolipid Degradation by Sphingolipid Activator Proteins and Anionic Lysosomal Lipids. Annu. Rev. Cell Dev. Biol. 2005, 21, 81–103, doi:10.1146/annurev.cellbio.21.122303.120013.
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500, doi:10.1038/emboj.2011.286.
- Perera, R.M.; Zoncu, R. The Lysosome as a Regulatory Hub. Annu. Rev. Cell Dev. Biol. 2016, 32, 223–253, doi:10.1146/annurev-cellbio-111315-125125.
- Settembre, C.; Fraldi, A.; Medina, D.L.; Ballabio, A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013, 14, 283–296, doi:10.1038/nrm3565.
- Lawrence, R.E.; Zoncu, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 2019, 21, 133–142, doi:10.1038/s41556-018-0244-7.
- Ishida, Y.; Nayak, S.; Mindell, J.A.; Grabe, M. A model of lysosomal pH regulation. J. Gen. Physiol. 2013, 141, 705–720, doi:10.1085/jgp.201210930.
- Zhao, J.; Benlekbir, S.; Rubinstein, J.L. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nat. Cell Biol. 2015, 521, 241–245, doi:10.1038/nature14365.
- Lim, C.-Y.; Zoncu, R. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 2016, 214, 653–664, doi:10.1083/jcb.201607005.
- Ferron, M.; Settembre, C.; Shimazu, J.; Lacombe, J.; Kato, S.; Rawlings, D.J.; Ballabio, A.; Karsenty, G. A RANKL-PKC -TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013, 27, 955–969, doi:10.1101/gad.213827.113.
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 Senses Lysosomal Amino Acids Through an Inside-Out Mechanism That Requires the Vacuolar H+-ATPase. Science 2011, 334, 678–683, doi:10.1126/science.1207056.
- Wullschleger, S.; Loewith, R.J.; Hall, M.N. TOR Signaling in Growth and Metabolism. Cell 2006, 124, 471–484, doi:10.1016/j.cell.2006.01.016.
- Hay, N.; Sonenberg, N. Upstream and Downstream of mTOR. Genes Dev. 2004, 18, 1926–1945, doi:10.1101/gad.1212704.
- Tee, A.R.; Blenis, J. mTOR, translational control and human disease. Semin. Cell Dev. Biol. 2005, 16, 29–37, doi:10.1016/j.semcdb.2004.11.005.
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976, doi:10.1016/j.cell.2017.02.004.
- Carroll, B.; A Dunlop, E. The lysosome: a crucial hub for AMPK and mTORC1 signalling. Biochem. J. 2017, 474, 1453–1466, doi:10.1042/bcj20160780.
- Zurli, V.; Montecchi, T.; Heilig, R.; Poschke, I.C.; Volkmar, M.; Wimmer, G.; Boncompagni, G.; Turacchio, G.; D’Elios, M.M.; Campoccia, G.; et al. Phosphoproteomics of CD2 signaling reveals AMPK-dependent regulation of lytic granule polariza-tion in cytotoxic T cells. Sci. Signal. 2020, 13, eaaz1965, doi:10.1126/scisignal.aaz1965.
- Wen, Z.; Jin, K.; Shen, Y.; Yang, Z.; Li, Y.; Wu, B.; Tian, L.; Shoor, S.; Roche, N.E.; Goronzy, J.J.; et al. N-myristoyltransferase deficiency impairs activation of kinase AMPK and promotes synovial tissue inflammation. Nat. Immunol. 2019, 20, 313–325, doi:10.1038/s41590-018-0296-7.
- Zhang, C.-S.; Jiang, B.; Li, M.; Zhu, M.; Peng, Y.; Zhang, Y.-L.; Wu, Y.-Q.; Li, T.Y.; Liang, Y.; Lu, Z.; et al. The Lysosomal v-ATPase-Ragulator Complex Is a Common Activator for AMPK and mTORC1, Acting as a Switch between Catabolism and Anabolism. Cell Metab. 2014, 20, 526–540, doi:10.1016/j.cmet.2014.06.014.
- González, A.; Hall, M.N.; Lin, S.-C.; Hardie, D.G. AMPK and TOR: The Yin and Yang of Cellular Nutrient Sensing and Growth Control. Cell Metab. 2020, 31, 472–492, doi:10.1016/j.cmet.2020.01.015.
- Christensen, K.A.; Myers, J.T.; Swanson, J.A. pH-dependent regulation of lysosomal calcium in macrophages. J. Cell Sci. 2002, 115, 599–607.
- Patel, S.; Cai, X. Evolution of acidic Ca2+ stores and their resident Ca2+-permeable channels. Cell Calcium 2015, 57, 222–230, doi:10.1016/j.ceca.2014.12.005.
- Clapham, D.E. Calcium Signaling. Cell 2007, 131, 1047–1058, doi:10.1016/j.cell.2007.11.028.
- Dong, X.-P.; Wang, X.; Xu, H. TRP channels of intracellular membranes. J. Neurochem. 2010, 113, 313–328, doi:10.1111/j.1471-4159.2010.06626.x.
- Luzio, J.P.; Bright, N.; Pryor, P. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem. Soc. Trans. 2007, 35, 1088–1091, doi:10.1042/bst0351088.
- Sun, X.; Yang, Y.; Zhong, X.Z.; Cao, Q.; Zhu, X.-H.; Zhu, X.; Dong, X.-P. A negative feedback regulation of MTORC1 activity by the lysosomal Ca2+ channel MCOLN1 (mucolipin 1) using a CALM (calmodulin)-dependent mechanism. Autophagy 2018, 14, 38–52, doi:10.1080/15548627.2017.1389822.
- Patel, S.; Docampo, R. Acidic calcium stores open for business: expanding the potential for intracellular Ca2+ signaling. Trends Cell Biol. 2010, 20, 277–286, doi:10.1016/j.tcb.2010.02.003.
- Morgan, A.; Platt, F.M.; Lloyd-Evans, E.; Galione, A. Molecular mechanisms of endolysosomal Ca2+ signalling in health and disease. Biochem. J. 2011, 439, 349–378, doi:10.1042/bj20110949.
- Shang, S.; Zhu, F.; Liu, B.; Chai, Z.; Wu, Q.; Hu, M.; Wang, Y.; Huang, R.; Zhang, X.; Wu, X.; et al. Intracellular TRPA1 medi-ates Ca2+ release from lysosomes in dorsal root ganglion neurons. J. Cell Biol. 2016, 215, 369–381, doi:10.1083/jcb.201603081.
- Tian, X.; Gala, U.; Zhang, Y.; Shang, W.; Jaiswal, S.N.; Di Ronza, A.; Jaiswal, M.; Yamamoto, S.; Sandoval, H.; DuRaine, L.; et al. A voltage-gated calcium channel regulates lysosomal fusion with endosomes and autophagosomes and is required for neuronal homeostasis. PLoS Biol. 2015, 13, e1002103, doi:10.1371/journal.pbio.1002103.
- Padamsey, Z.; McGuinness, L.; Bardo, S.J.; Reinhart, M.; Tong, R.; Hedegaard, A.; Hart, M.L.; Emptage, N.J. Activi-ty-Dependent Exocytosis of Lysosomes Regulates the Structural Plasticity of Dendritic Spines. Neuron 2017, 93, 132–146, doi:10.1016/j.neuron.2016.11.013.
- Venkatachalam, K.; Montell, C. TRP Channels. Annu. Rev. Biochem. 2007, 76, 387–417, doi:10.1146/annurev.biochem.75.103004.142819.
- Puertollano, R.; Kiselyov, K. TRPMLs: in sickness and in health. Am. J. Physiol. Physiol. 2009, 296, F1245–F1254, doi:10.1152/ajprenal.90522.2008.
- Cheng, X.; Shen, D.; Samie, M.; Xu, H. Mucolipins: Intracellular TRPML1-3 channels. FEBS Lett. 2010, 584, 2013–2021, doi:10.1016/j.febslet.2009.12.056.
- Dong, X.-P.; Shen, D.; Wang, X.; Dawson, T.; Li, X.; Zhang, Q.; Cheng, X.; Zhang, Y.; Weisman, L.S.; Delling, M.; et al. PI(3,5)P2 controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat. Commun. 2010, 1, 1–11, doi:10.1038/ncomms1037.
- Sun, M.; Goldin, E.; Stahl, S.; Falardeau, J.L.; Kennedy, J.C.; Jr, J.S.A.; Bove, C.; Kaneski, C.R.; Nagle, J.; Bromley, M.C.; et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 2000, 9, 2471–2478, doi:10.1093/hmg/9.17.2471.
- Bargal, R.; Avidan, N.; Ben-Asher, E.; Olender, Z.; Zeigler, M.; Frumkin, A.; Raas-Rothschild, A.; Glusman, G.; Lancet, D.; Bach, G. Identification of the gene causing mucolipidosis type IV. Nat. Genet. 2000, 26, 118–122, doi:10.1038/79095.
- Dong, X.-P.; Cheng, X.; Mills, E.; Delling, M.; Wang, F.; Kurz, T.; Xu, H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nat. Cell Biol. 2008, 455, 992–996, doi:10.1038/nature07311.
- Dong, X.-P.; Wang, X.; Shen, D.; Chen, S.; Liu, M.; Wang, Y.; Mills, E.; Cheng, X.; Delling, M.; Xu, H. Activating Mutations of the TRPML1 Channel Revealed by Proline-scanning Mutagenesis. J. Biol. Chem. 2009, 284, 32040–32052, doi:10.1074/jbc.m109.037184.
- Medina, D.L.; Fraldi, A.; Bouche, V.; Annunziata, F.; Mansueto, G.; Spampanato, C.; Puri, C.; Pignata, A.; Martina, J.A.; Sardi-ello, M.; et al. Transcriptional Activation of Lysosomal Exocytosis Promotes Cellular Clearance. Dev. Cell 2011, 21, 421–430, doi:10.1016/j.devcel.2011.07.016.
- Samie, M.; Wang, X.; Zhang, X.; Goschka, A.; Li, X.; Cheng, X.; Gregg, E.; Azar, M.; Zhuo, Y.; Garrity, A.G.; et al. A TRP Channel in the Lysosome Regulates Large Particle Phagocytosis via Focal Exocytosis. Dev. Cell 2013, 26, 511–524, doi:10.1016/j.devcel.2013.08.003.
- Bargal, R.; Avidan, N.; Olender, T.; Ben Asher, E.; Zeigler, M.; Raas-Rothschild, A.; Frumkin, A.; Ben-Yoseph, O.; Friedlender, Y.; Lancet, D.; et al. Mucolipidosis type IV: NovelMCOLN1 mutations in Jewish and non-Jewish patients and the frequency of the disease in the Ashkenazi Jewish population. Hum. Mutat. 2001, 17, 397–402, doi:10.1002/humu.1115.
- Shen, D.; Wang, X.; Li, X.; Zhang, X.; Yao, Z.; Dibble, S.; Dong, X.-P.; Yu, T.; Lieberman, A.P.; Showalter, H.D.; et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 2012, 3, 731, doi:10.1038/ncomms1735.
- Chen, C.-C.; Butz, E.S.; Chao, Y.-K.; Grishchuk, Y.; Becker, L.; Heller, S.; Slaugenhaupt, S.A.; Biel, M.; Wahl-Schott, C.; Grimm, C. Small Molecules for Early Endosome-Specific Patch Clamping. Cell Chem. Biol. 2017, 24, 907–916.e4, doi:10.1016/j.chembiol.2017.05.025.
- Plesch, E.; Chen, C.-C.; Butz, E.S.; Rosato, A.S.; Krogsaeter, E.; Yinan, H.; Bartel, K.; Keller, M.; Robaa, D.; Teupser, D.; et al. Selective agonist of TRPML2 reveals direct role in chemokine release from innate immune cells. eLife 2018, 7, 39720-, doi:10.7554/eLife.39720.
- Xu, H.; Delling, M.; Li, L.; Dong, X.; Clapham, D.E. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc. Natl. Acad. Sci. USA 2007, 104, 18321–18326.
- Samie, M.A.; Grimm, C.; Evans, J.A.; Curcio-Morelli, C.; Heller, S.; Slaugenhaupt, S.A.; Cuajungco, M.P. The tissue-specific expression of TRPML2 (MCOLN-2) gene is influenced by the presence of TRPML1. Pflügers Archiv 2009, 459, 79–91, doi:10.1007/s00424-009-0716-5.
- Cuajungco, M.P.; Da Silva, J.F.M.; Habibi, A.; Valadez, J.A. The mucolipin-2 (TRPML2) ion channel: a tissue-specific protein crucial to normal cell function. Pflügers Archiv 2016, 468, 177–192, doi:10.1007/s00424-015-1732-2.
- Sun, L.; Hua, Y.; Vergarajauregui, S.; Diab, H.I.; Puertollano, R. Novel Role of TRPML2 in the Regulation of the Innate Im-mune Response. J. Immunol. 2015, 195, 4922–4932, doi:10.4049/jimmunol.1500163.
- Karacsonyi, C.; Miguel, A.S.; Puertollano, R. Mucolipin-2 Localizes to the Arf6-Associated Pathway and Regulates Recycling of GPI-APs. Traffic 2007, 8, 1404–1414, doi:10.1111/j.1600-0854.2007.00619.x.
- Chen, C.-C.; Krogsaeter, E.; Butz, E.S.; Li, Y.; Puertollano, R.; Wahl-Schott, C.; Biel, M.; Grimm, C. TRPML2 is an os-mo/mechanosensitive cation channel in endolysosomal organelles. Sci. Adv. 2020, 6, eabb5064, doi:10.1126/sciadv.abb5064.
- Kim, S.W.; Kim, D.H.; Park, K.S.; Kim, M.K.; Park, Y.M.; Muallem, S.; So, I.; Kim, H.J. Palmitoylation controls trafficking of the intracellular Ca2+ channel MCOLN3/TRPML3 to regulate autophagy. Autophagy 2019, 15, 327–340, doi:10.1080/15548627.2018.1518671.
- Kim, H.J.; Soyombo, A.A.; Tjon-Kon-Sang, S.; So, I.; Muallem, S. The Ca2+Channel TRPML3 Regulates Membrane Traffick-ing and Autophagy. Traffic 2009, 10, 1157–1167, doi:10.1111/j.1600-0854.2009.00924.x.
- Martina, J.A.; Lelouvier, B.; Puertollano, R. The Calcium Channel Mucolipin-3 is a Novel Regulator of Trafficking Along the Endosomal Pathway. Traffic 2009, 10, 1143–1156, doi:10.1111/j.1600-0854.2009.00935.x.
- Castiglioni, A.J.; Remis, N.N.; Flores, E.N.; García-Añoveros, J. Expression and Vesicular Localization of Mouse Trpml3 in Stria Vascularis, Hair Cells, and Vomeronasal and Olfactory Receptor Neurons. J. Comp. Neurol. 2011, 519, 1095–1114, doi:10.1002/cne.22554.
- Remis, N.N.; Wiwatpanit, T.; Castiglioni, A.J.; Flores, E.N.; Cantú, J.A.; García-Añoveros, J. Mucolipin Co-deficiency Causes Accelerated Endolysosomal Vacuolation of Enterocytes and Failure-to-Thrive from Birth to Weaning. PLoS Genet. 2014, 10, e1004833, doi:10.1371/journal.pgen.1004833.
- Kim, H.J.; Li, Q.; Tjon-Kon-Sang, S.; So, I.; Kiselyov, K.; Soyombo, A.A.; Muallem, S. A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 2008, 27, 1197–1205, doi:10.1038/emboj.2008.56.
- Di Palma, F.; Belyantseva, I.A.; Kim, H.J.; Vogt, T.F.; Kachar, B.; Noben-Trauth, K. Mutations in Mcoln3 associated with deafness and pigmentation defects in varitint-waddler (Va) mice. Proc. Natl. Acad. Sci. USA 2002, 99, 14994–14999.
- Piao, S.; Amaravadi, R.K. Targeting the lysosome in cancer. Ann. N. Y. Acad. Sci. 2016, 1371, 45–54, doi:10.1111/nyas.12953.
- Davidson, S.M.; Heiden, M.G.V. Critical Functions of the Lysosome in Cancer Biology. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 481–507, doi:10.1146/annurev-pharmtox-010715-103101.
- Hämälistö, S.; Jäättelä, M. Lysosomes in cancer—living on the edge (of the cell). Curr. Opin. Cell Biol. 2016, 39, 69–76, doi:10.1016/j.ceb.2016.02.009.
- Fehrenbacher, N.; Jäättelä, M. Lysosomes as Targets for Cancer Therapy: Figure 1. Cancer Res. 2005, 65, 2993–2995, doi:10.1158/0008-5472.can-05-0476.
