Riboswitches reside in the 5'- untranslated region of RNA and regulate genes involved in the homeostasis of molecules or Ions they bind to. Since their discovery at the beginning of this century, some of the riboswitches have been regarded as potential antibacterial targets due to their role in the regulation of the homeostasis of the essential metabolites. The pharmacological potential of such riboswitches is being explored u various approaches which include fragment screening, high-throughput screening and rational ligand design guided by X-ray crystallography.
- antibacterial drug target
- riboswitch
- structure-based drug design
- fragment screening
- high-throughput screening
1. Introduction
The emergence of bacterial resistance to antibiotics has become an urgent and serious threat to global public health. Resistant bacteria are expected to cause nearly 10 million deaths each year globally by 2050 [1]. As many pathogenic bacteria are evolving to persist against all existing antibiotics, it is feared that the health system will become incapacitated against serious bacterial infections [2][3][2,3]. Indeed, to address this concern, the World Health Organization (WHO) has released a priority list of bacterial pathogens that need immediate attention, to focus antibiotic drug discovery efforts [4]. Consequently, a need for a new generation of antibiotics with novel mechanisms of action against resistant bacteria is increasingly being recognized [5] [5] and current drug discovery programs are exploring both proteins, as well as nucleic acid targets [6][7][6,7].
Riboswitches are a novel antibacterial drug target class that could deliver urgently needed antibiotics via a new mechanism of action. They occur almost exclusively in bacteria and regulate the biosynthesis and transport of amino acids and essential metabolites, such as coenzymes, nucleobases and their derivatives by binding small molecules [8][9][8,9]. Residing in the 5′ untranslated region (UTR) of mRNA, these cis-regulatory elements are structured non-coding RNAs that adopt alternative 3D-conformations to function as genetic switches (“ribo-switching”) [10][11][12][10,11,12]. Riboswitches are comprised of two domains; an aptamer domain that selectively binds the cognate ligands and an expression platform that translates the presence of ligands into expression or repression of downstream genes (Figure 1). When the concentration of a cognate ligand in the intracellular milieu increases beyond a threshold, its binding to the aptamer domain induces a conformational change. In most cases, this ligand-induced alternate conformation leads to transcriptional attenuation by the formation of a terminator or inhibition of translation by sequestering the ribosomal binding site (RBS) or both, followed by switching off the downstream gene or operon [13][14][15][13,14,15]. These riboswitch elements are termed “OFF-switches” (Figure 1a). In other cases, the ligand-induced alternate conformation leads to the formation of an anti-terminator stem and/or release of the RBS from the terminator stem followed by activation of the downstream target gene(s). Accordingly, these are referred to as “ON-switches” (Figure 1b).

Figure 1.
a
b
a
b
a
b
Riboswitches are widespread in bacteria. Currently, 28 experimentally validated riboswitch classes are known, which are distributed across more than 6000 bacterial species [16][17][16,17]. Riboswitches are categorized based on their cognate ligand and the fold of their aptamer domain . Biochemically, these natural ligands span a wide spectrum of biomolecules, namely coenzymes, nucleotides and their derivatives, ions, amino acids, phosphorylated sugars and guanidine. Some riboswitches are widely present across human bacterial pathogens and therefore, are suitable targets for broad-spectrum antibiotics, while less widespread riboswitches are potential targets for selective antibacterial drugs. The thiamine pyrophosphate (TPP) riboswitch is the most prevalent riboswitch and is the only one present in eukaryotes, including algae [18], fungi [19][20][21] [19,20,21] and plants [22][23][22,23]. Other riboswitches prevalently present across human bacterial pathogens (>20) include the flavin mononucleotide (FMN), cobalamin, glutamine-fructose-6-phosphate (glmS), lysine, S-adenosyl methionine-I (SAM-I) and glycine riboswitches.
Riboswitches bind to their cognate ligand mostly with high affinity (KD < 10 nM) and discriminate strongly against related compounds (drop in affinity by around two orders of magnitude) [24][25][26][25,26,27]. This suggests the possibility to rationally design potent structural or functional analogues of the natural ligands that can be exploited to starve bacterial cells of essential metabolites and thereby can function as novel antibacterial drugs. To harvest the full potential of riboswitches as antibacterial targets, detailed structural and biochemical knowledge is crucial. The sustained interest in the characterization of various riboswitches has led to a gradual improvement in riboswitch structure wealth over time. Currently, more than 300 crystal structures of riboswitches are available in the Protein Databank (PDB) and at least one representative of most riboswitch classes has been characterized. This has revealed an architectural diversity among different riboswitch classes, leading to a broad categorization into two groups: pseudoknotted and junctional riboswitches [27][28][29][30][28,29,30,31]. Structural principles of selective binding to distinct riboswitches are now well understood which has in some cases contributed to the rational design of novel ligands [31][32][33][32,33,34].
In order to discriminate cognate ligands from related compounds in vivo, riboswitches have evolved to form a binding pocket that specifically recognizes their natural ligands. In fact, different riboswitches employ distinct strategies to achieve a high affinity and an appreciable degree of selectivity. Some riboswitches, like the lysine and purine riboswitches, possess binding pockets with perfect shape complementarity to cognate ligands and encapsulate them almost entirely and thus exclude binding of related compounds [34][35][35,36]. Often, hydrogen bonding with the ligand is an important means of achieving high specificity. This is exemplified by changing the specificity of the adenine riboswitch from adenine to guanine by mutating U74 in the binding pocket to C74 and thus requiring a different ligand to form a Watson–Crick base pair [36][37]. In addition to nucleobases, also the ribose and phosphate groups of the nucleotides are involved in recognizing the ligands, albeit this varies depending on the riboswitch. The interactions with the negatively charged group of ligands (e.g., a phosphate group in the case of TPP and FMN) are often mediated by cations, such as divalent Mg+2 and monovalent K+, either directly through coordinated bonds or indirectly through water molecule(s) [37][38][38,39]. Positively charged groups of ligands, such as those in lysine or SAM, typically interact directly with the negative phosphate backbone and other surrounding polar but neutral atoms of the base or sugar moieties [34][39][35,40]. Pi–pi interactions between aromatic rings of the ligands and nucleobases are another driving factor for ligand binding. Altogether, the diversity of interaction patterns together with the shape diversity of the binding pockets allow the different riboswitches to bind diverse ligands with high specificity.
From a drug discovery point of view, not all riboswitches are of equal interest. First of all, in order to be pharmacologically relevant, the riboswitch needs to be present in pathogenic bacteria, control the expression of the essential gene(s) and act as an off-switch. Riboswitches that occur in a range of pathogenetic bacteria offer the opportunity to develop broad-spectrum antibiotics while riboswitches that are not widely spread can be suitable targets for narrow-spectrum antibiotics that are sought after to avoid detrimental effects on the microbiome and to lower the risk of spreading resistance [40][41]. Second, the binding site of the riboswitch needs to have a pocket that is suitable to bind a drug-like ligand with high affinity. While some binding sites are too small, too polar or too solvent exposed to be suited, others have properties that render them suitable candidates for drug discovery efforts [41][42][43][42,43,44].
2. TPP Riboswitch Ligands
The TPP riboswitch is the most abundant riboswitch. This riboswitch regulates the expression of genes involved in the synthesis and transport of TPP (Figure 2a) [16][17][44][45] [16,17,45,46] which is an essential cofactor for the carboxylation and decarboxylation of various metabolic intermediates in the carbohydrate and amino-acid metabolism [46][47]. The TPP riboswitch is probably the most diverse riboswitch with respect to the mechanisms for regulating gene expression. These include modulating mRNA decay, Rho dependent transcription termination, alternate splicing, transcription termination through the formation of an intrinsic terminator and translation inhibition through sequestration of ribosomal binding sites (RBS) [14][47][14,48]. This off-switch is present in 48 human pathogens [48][49], including most of the pathogens of the WHO priority list . Thus, this riboswitch is one of the most suitable candidates for broad spectrum antibiotics.
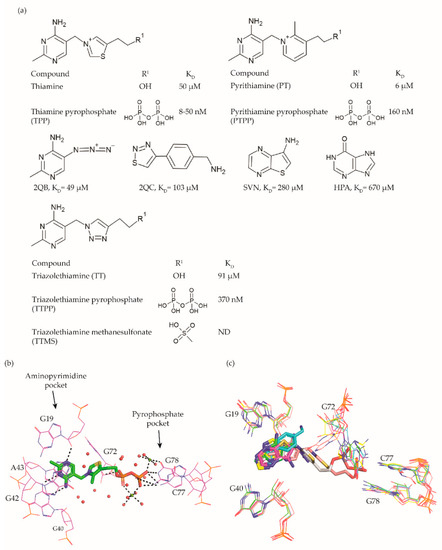
Figure 2.
a) Chemical structures and binding affinities of ligands taken from various studies [31][44][49][50]. (
b
c
A compound that established the proof of principle for the TPP riboswitch as an antibacterial drug target became available well before the riboswitch was discovered. Pyrithiamine (PT) (Figure 2a), originally designed and synthesized as a structural analogue of thiamine to study thiamine metabolism, is toxic to fungi as well as bacteria [44][51][52][45,50,51]. PT becomes phosphorylated intracellularly to form pyrithiamine pyrophosphate (PTPP) (Figure 2a). PTPP has a similar binding affinity to the B. subtilis TPP riboswitch as TPP in vitro (160 nM vs. 50 nM) and turns off the expression of downstream gene(s) involved in TPP biosynthesis or transport. Further, bacteria resistant to PT were found to have mutations predominantly in the genetic region coding for the TPP riboswitch, hinting that the TPP riboswitch is the target for this compound.
Meanwhile, crystal structures of the TPP riboswitch aptamer domain in complex with TPP as well as other ligands were reported at a resolution ranging from 2.05 Å to 3.2 Å [25][31][37][53][26,32,38,54]. These structures have played a crucial role in revealing the binding modes of the ligands and for suggesting strategies to explore structure–activity relationships (SAR). TPP binds to the riboswitch in an extended conformation (Figure 2b). The aminopyrimidine moiety of TPP is sandwiched between the bases of G42 and A43 and forms hydrogen bonds with polar entities of G40 and G19. The pyrophosphate moiety of TPP is coordinated to two Mg+2 ions and also forms direct hydrogen bonds with C77 and G78. The hexa-coordinated Mg+2 ions in this pocket are positioned by direct and water-mediated hydrogen bonds with the surrounding residues and thereby facilitate the binding of the negatively charged phosphate group. The thiazole group appears to be less constrained and only forms hydrophobic contacts with G72. The nucleotides in close contact with TPP are highly conserved across TPP riboswitch sequences, suggesting that potential ligands will bind to all TPP riboswitch binding sites.
Initial efforts to identify novel TPP riboswitch binders were made using a fragment-based approach. Using [3H]thiamine dependent equilibrium dialysis, Cressina et al. screened 1300 fragments for their ability to displace a radioligand [54] [55]. A total of 20 hits were identified in the initial screen. However, further validation using a combination of NMR spectroscopy, isothermal titration calorimetry (ITC) measurements and counter-screening against the lysine responsive (lysC) aptamer, the hits were reduced to 17 structurally diverse compounds with dissociation constants in the range of 22–670 μM. None of these fragments were able to reduce the expression of a luciferase gene used as a reporter in an in vitro transcription termination assay (IVTT). The failure to suppress the expression of a downstream reporter gene upon small molecule binding could either be caused by the fragments not being able to induce the conformational change essential for switching off the expression or by the fragments not binding strong enough to the longer transcript used in the IVTT assay.
As a first step towards the rational elaboration of the fragment hits, Warner et al. determined crystal structure of four fragment hits (2QB, 2QC, SVN, and HPA, Figure 2a) bound to the TPP riboswitch [31][32]. Occupying the pocket within the aminopyrimidine sensor helix of the TPP aptamer, each of these fragments was found to induce a conformational change in G72, while keeping the overall conformation of the aptamer similar to that of the TPP bound form (Figure 2c). The determined structures are valuable tools to facilitate structure-guided fragment elaboration to increase their binding affinities.
Alternatively, structural analogues of TPP have also been developed. By systematically replacing the two rings and the pyrophosphate tail of TPP with analogues, Chen et al. established the SAR for these compounds (Figure 2a) [49][52]. The replacement of the aminopyrimidine ring with various heterocyclic rings significantly compromised the binding affinity. However, the replacement of the thiazolium ring with a related 5-member heterocyclic ring only modestly affected the affinity. In fact, positively charged heterocycles are well tolerated in this position whereas, replacing the thiazole ring of TPP with open-chain analogues severely compromised the binding affinity, suggesting that a ring structure in this position is important. As expected, the negative charge of the pyrophosphate tail is vital to attain high affinity as replacing the tri-anionic group with bi- or mono- anionic groups reduced the binding affinity by at least two and four orders of magnitude, respectively. Additionally, the pKa of the tri-anionic group is also crucial as the replacement of the pyrophosphate group (pKa ~5.77) with a methylenediphosphonate (pKa ~7.45) improved the affinity by a magnitude of two orders. Encouragingly, ligand affinity and the riboswitch-mediated repression of the reporter gene appeared positively correlated. TPP analogues with the affinity comparable to that of TPP repressed the gene expression to the same extent as TPP, whereas those with mediocre affinity repressed the reporter gene to an extent similar to the weaker ligands. Lünse et al. further investigated the binding of the TTP riboswitch to triazolethiamine (TT) (Figure 2a) and a series of its derivatives using an intracellular gene reporter assay [50][53]. TT was still the most potent compound with an IC50 of 91 μM whereas, that of its derivatives range from 20 µM to the high micromolar. In contrast to the TT, whose potency is dependent on active transport and an in vivo phosphorylation, its methanesulfonate analogue (TTMS) is interesting despite with weaker IC50 value as it appears to function independently of intracellular enzymes (Figure 2a).
Altogether, these studies demonstrate that only a few modifications of TPP are allowed to maintain high affinity. Further, phospho-mimicking compounds could be an alternative approach to develop TPP riboswitch dependent effective antibacterial agents. Finally, the discovered fragment hits offer opportunities to develop ligands that are dissimilar to TPP.
3. FMN Riboswitch Ligands
The FMN riboswitch regulates the expression of genes involved in biosynthesis and transport of riboflavin (vitamin B12, Figure 3a) [55][56][56,57]. As a precursor of the coenzymes FMN (Figure 3a and flavin adenine dinucleotide (FAD), riboflavin plays an essential role in oxidative phosphorylation, β-oxidation and the Kreb’s cycle [57][58][59][58,59,60]. Although riboflavin and FAD also bind to this OFF riboswitch, only FMN associates with high enough affinity to repress the expression of the downstream gene(s) either by transcriptional termination or by translation inhibition. The FMN riboswitch is the third most widespread riboswitch known to date and is found in 41 human pathogens, including seven from the WHO priority pathogen list [13][17][48][13,17,49]. In brief, its essential role in regulating bacterial physiology, the high conservation of the ligand binding domain, the absence of a counterpart in humans and its high abundance make the FMN riboswitch a promising target for broad-spectrum antibacterials.
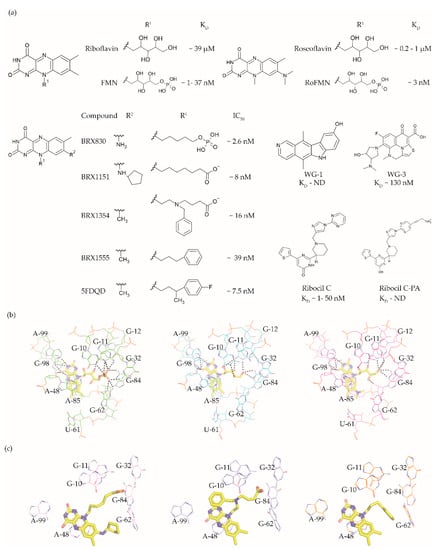
Figure 3.
a
D
50 values taken from various studies [33][60][61][62][63]. (
b
c
The FMN riboswitch is an established target for the antibacterial compound roseoflavin (Figure 3a). Roseoflavin (Figure 3a) is a natural riboflavin analogue synthesized by Streptomyces davawensis [64][65][65,66]. In its phosphorylated form (RoFMN), roseoflavin inhibits the growth of Bacillus subtilis, Escherichia coli, and Listeria monocytogenes by targeting the FMN riboswitch [64][66][67][65,67,68]. This leads to the suppression of riboflavin biosynthesis and transport, and thus the cells are starved of riboflavin. RoFMN binds to the FMN aptamer domain with a similar affinity as FMN (< 5 nM), resulting in the repression of the regulated genes [68][69][69,70]. As RoFMN is structurally highly related to FMN, it also binds to flavoenzymes. Therefore, the antibacterial effect of RoFMN is mediated by both the FMN riboswitch and flavoenzymes. Indeed, in roseoflavin-resistant mutants, mutations are found in the FMN riboswitch aptamer domain and flavoenzymes [38][61][39,62]. Despite RoFMN not being selective, its FMN riboswitch-mediated antibiotic effect underscores the promise of this riboswitch for antibacterial drug discovery.
The crystal structures of the FMN riboswitch in complex with FMN analogues have been determined with resolutions of around 2.9 to 3.45 Å. These structures revealed that the isoalloxazine rings of the ligands form similar interactions with the aptamer domain. In the complex with roseoflavin, U61 only modestly reorients in order to accommodate the relatively bulkier dimethylamino group in of ligand (Figure 3b) [38][39]. In contrast, differently substituted alkyl chains in the R1 position adopt different conformations in the various structures (Figure 3c). In fact, the ribityl moiety was found to have minimal contribution to the affinity and is discussed in detail in the following paragraph. In the case of FMN, the terminal phosphate group interacts with the aptamer domain through hydrogen bonds directly as well as indirectly through Mg+2 ion. These interactions appear to be crucial for binding as riboflavin and roseoflavin, which lack the phosphate group, have a 1000 and 100-fold lower affinity, respectively [38][61][62][39,62,63]. In contrast to the TPP riboswitch, which undergoes a considerable conformational change during ligand binding, various structural and chemical probing methods show that in the FMN riboswitch, the ligand binding pocket adopts a similar conformation in both the absence and presence of FMN [38][62][70][71][39,63,71,72]. Altogether, the ligand binding region of the FMN riboswitch offers a balance of hydrophobic and hydrophilic interactions and has been predicted to bind drug-like ligands using computational methods [41][42][43][42,43,44].
Recently, a few promising small molecules with antibiotic activity have been discovered to selectively target the FMN riboswitch using three different approaches: a structure-based approach, a phenotypic screen, and high-throughput screening (HTS).
Vicens et al. employed a structure-based approach to identify RoFMN analogues that would not require intracellular phosphorylation for antibacterial activity [33][34]. Simplifying the ribityl group of RoFMN to an alkyl chain but maintaining the negative charge at its end led to the compounds BRX830 (IC50 ~2.6 nM) and BRX1151 (IC50 ~8 nM) with similar inhibitory activity as RoFMN (IC50 ~3 nM) in an in vitro transcription assay (Figure 3a). A further modification to contain an aromatic side chain led to the compounds BRX1354 (IC50 ~16 nM) and BRX1555 (IC50 ~39 nM), and eventually the drug-like molecule 5FDQD (Figure 3a). The latter no longer requires a negative charge for potent inhibition of the transcription reaction (IC50 ~7.5 nM). Further, analysis through X-ray crystallography showed that the compounds BRX1151, BRX1354 and BRX1555 bind to the aptamer domain in a similar fashion as FMN, with their isoalloxazine ring intercalated between A48 and the A85-G98 pair and hydrogen bonding with A99 (Figure 3c). Interestingly, the binding pocket where the FMN ribityl side chain is located is rather promiscuous as a variety of functional groups on the alkyl chain at R1 could be accommodated between G10, G11 and G62 (Figure 3c). Further, the discovery of these compounds clearly demonstrates that the negatively charged chain of FMN could be replaced with hydrophobic moieties without compromising affinity, resulting in potent drug-like ligands. An in vitro characterization of 5FDQD revealed potent and rapid bactericidal activity against Clostridium difficile with only a modest effect on the culturable cecal flora of mice [72][73]. In a mouse model of C. difficile infection, the antibacterial performance of 5FDQD is similar to that of fidaxomicin and vancomycin until 8 days post-infection. Despite the promising results of 5FDQD, it is noteworthy that its definitive mode of action remains elusive as no resistant mutants could be obtained to confirm the target.
Another example of a potent FMN riboswitch ligand is ribocil (KD = 13 nM, Figure 3A), discovered serendipitously through a phenotypic screen [60][61]. The screening involved testing a library of ~57,000 synthetic compounds against an E. coli strain defective in lipopolysaccharide synthesis and drug efflux (MB5746). The only hit selective to riboflavin synthesis in the screen was ribocil. Characterization of the ribocil resistant mutants mapped all base pair changes to the FMN riboswitch upstream of the ribB gene (involved in riboflavin biosynthesis), establishing it as the sole target. Ribocil is a racemic mixture of the R- (ribocil-A) and S-enantiomer (ribocil-B), and it is the latter that predominantly inhibits the riboflavin biosynthesis and consequently bacterial growth. A further structural elaboration led to the compound ribocil-C (Figure 3a), which has approximately eight-fold higher potency against the Gram-positive bacteria E. faecalis and methicillin-resistant S. aureus [73][74][74,75]. Interestingly, in a murine E. coli (MB5746) septicaemia model of infection, ribocil-C demonstrated a dose-dependent reduction in the bacterial burden without any evident toxic side effects. Recently, Motika et al. modified ribocil-C, which lacks activity against Gram-negative pathogens, to ribocil C-PA (Figure 3a), which is also active against Gram-negative bacteria [63][64]. In fact, ribocil C-PA inhibited all multi-drug resistant clinical strains of E. coli and K. pneumonia tested in the study. Ribocil C-PA is 16-fold more potent against strains of E. coli (MIC = 4 μg/mL), Enterobacter cloacae (MIC) = 4 μg/mL), and K. pneumoniae (MIC = 4 μg/mL) than ribocil C (minimum inhibitory concentration MIC > 64 μg/mL). Encouragingly, ribocil C-PA was also effective in mice in E. coli infection models for acute pneumonia and sepsis. Indeed, ribocil_C-PA rescued 80% of mice infected with pathogenic strains of E. coli, whereas ribocil-C was ineffective. Unfortunately, both ribocil C and ribocil C-PA led to a high frequency of resistance (FOR) in E. coli strains (at the order of 10−6).
More recently, Rizvi et al. demonstrated the application of the affinity selection mass spectrometry (AS-MS) based automated ligand detection system (ALIS) for the discovery of selective riboswitch ligands. They used a HTS approach to screen a library of ~53,000 unbiased small molecules against the FMN riboswitch [75][76]. Although the hit compounds WG-1 (KD not determined, Figure 3a) and WG-3 (KD ~130 nM, Figure 3a) bind to the FMN riboswitch in an FMN competitive manner, both exhibited riboflavin independent antibacterial activity against E. coli (MB5746). This suggests that both are non-selective in terms of their mode of action, and therefore lack potential as FMN riboswitch targeting antibacterials.
Altogether, these studies demonstrate the potential of the FMN riboswitch as a promising target for the next generation of antibiotics, however, high FOR associated with the FMN riboswitch ligands remains a serious concern.
4. glmS Riboswitch Ligands
The glmS riboswitch is unique as it is a self-cleaving ribozyme that uses the cognate ligand—glucosamine-6-phosphate (GlcN6P, Figure 4a)—as a coenzyme to catalyse the cleavage reaction. The riboswitch is predominantly present in Gram-positive bacteria and resides in the 5′-UTR of the glutamine-fructose-6-phosphate aminotransferase (glmS) gene, which catalyses the synthesis of GlcN6P [76][77][77,78]. At micromolar concentration, GlcN6P induces glmS riboswitch self-cleavage to produce an upstream fragment with a 2′,3′-cyclic phosphate (2′,3′-cP) end and a downstream fragment with a 5′-OH moiety (Figure 4b) [78][79][79,80]. The latter is rapidly degraded by RNase J1 in B. subtilis, suggesting that the glmS riboswitch affects the mRNA stability in a GlcN6P dependent manner [80][81]. Notably, the catalytic core of the riboswitch is conserved across bacterial species (>97% identity), which allows it to function in the same way despite sequence and structure variability outside of the binding pocket. As GlcN6P is an essential metabolite for normal growth [81][82] [82,83] and an essential precursor for the synthesis of the bacterial cell wall [83][84], the glmS riboswitch is regarded as a promising target to develop antibacterial drugs against various bacterial pathogens .
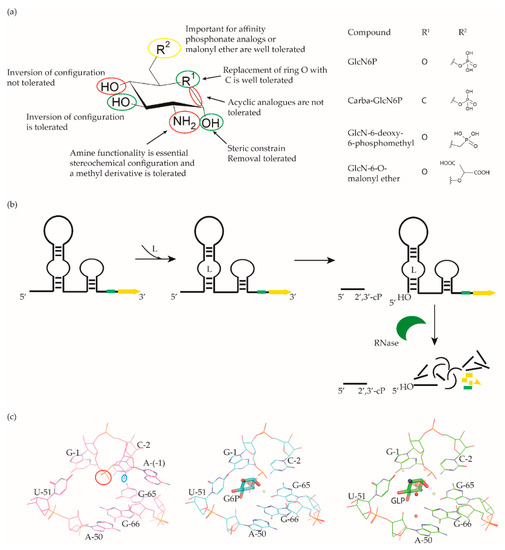
Figure 4.
glmS
a
b
glmS
glmS
c
glmS
McCarthy et al. were the first to characterize the ligand requirements to induce glmS riboswitch self-cleavage using a series of amine-containing analogues. They revealed that the amine functionality of GlcN6P is essential for catalysis [84][85]. This observation was also corroborated by various structural studies [85][86][87][88][86,87,88,89]. Effectively, there is no conformational or positional rearrangement of the active site upon binding the ribozyme inhibitor glucose-6-phosphate (Glc6P) or GlcN6P (Figure 4c). Further, comparison of the apo form with the Glc6P or GlcN6P bound glmS riboswitch structures showed that the ligands bind to a preorganized active site wherein the nucleophile for the catalysis (2′-OH group of A(-1)) and the leaving group (5′-phosphate group of G1) are in close proximity (Figure 4c). Consequently, binding of GlcN6P activates the riboswitch through the positioning of its primary amine close to the 5′-O of G1 to function as a general acid accelerating the cleavage reaction. It is important to note that replacing the phosphate group of Glc6NP with a sulphate group reduced the affinity by 100-fold, indicating that the phosphate group is critical for binding.
Up to now, efforts to explore the glmS riboswitch as an antibacterial drug target are focused on hit discovery and SAR studies. Evaluation of the discovered compounds against pathogenic bacteria is still outstanding. The Breaker group developed a fluorescence resonance energy transfer (FRET) based high-throughput assay to screen structural and stereochemical analogues of GlcN6P and a library of 960 compounds approved for use in humans [89][90][90,91]. Although no hit was identified from the latter, screening of Glc6NP analogues revealed the importance of various functional groups to induce glmS ribozyme activity summarized below (Figure 4a). Using a fluorescence polarization-based assay Mayer et al. screened a library of > 5000 compounds which follow Lipinski’s rule of five and novel analogues of GlcN6P. The latter included various 1-deoxy, 1-methyl glycoside, and carba-derivatives of Glc6NP. None of the compounds from the diverse library and none of the 1-deoxy and 1-methyl glycoside derivatives, which also lacked one or more equatorial hydroxyl moieties were active. In contrast, the carba-analogue of Glc6NP efficiently induced the self-cleavage of the glmS riboswitch, suggesting that the ring oxygen is dispensable for the ribozyme activity (Figure 4a) [91][92][92,93]. More recently, the group explored mono-fluorinated carba variant of GlcN6P [93][94]. The most effective compound from the study- fluoro-carba-a-D-GlcN6P (Figure 4a), had a significantly lower activity compared to its non-fluorinated analogue (EC50 = 300 μM vs. 6.2 μM), rendering this compound series less promising. In addition, a series of phosphate analogues of GlcN6P were also designed and analysed for their ability to induce glmS ribozyme activity [94][95]. Among these, 6-deoxy-6-phosphonomethyl and 6-O-malonyl ether analogues (Figure 4a) demonstrated potent ribozyme activity.
Taking all studies together, a comprehensive SAR of glmS riboswitch activation has emerged. The ring oxygen is non-essential, whereas OH groups are important for the activity as they interact with RNA through hydrogen binding (Figure 4b). Although, due to inconsistent data, it is difficult to compare the affinity of potent analogues discovered through different studies, carba-GlcN6P and 6-deoxy-6-phosphonomethyl and 6-O-malonyl-ether analogues of GlcN6P appear to be the most promising compounds.
Notably, none of these compounds are yet tested for their antibacterial effect or target selectivity. All discovered glmS riboswitch activators are GlcN6P-analogs, and screening of diverse libraries did not reveal any hits. However, these libraries were rather small. Perhaps, more hits could be found by screening larger and more diverse compound collections. Conclusively, the feasibility of developing antibacterial agents by targeting glmS riboswitch can largely be considered under-explored.
5. Guanine Riboswitch Ligands
The guanine riboswitch resides in the 5′-UTR of genes involved in the transport and biosynthesis of purine nucleotides [24][25]. This riboswitch is moderately widespread and present in two of the WHO priority pathogens; Staphylococcus aureus and Streptococcus pneumoniae [16][17][16,17]. Disrupting guanine riboswitch regulated gene expression leads to a compromised growth rate of B. subtilis [95][96]. Further, attempts to generate a B. subtilis strain that lacks all transcriptional units under the regulation of the guanine riboswitch failed. Altogether, these experiments suggest that the guanine riboswitch is a suitable target for narrow-spectrum antibiotics.
The guanine riboswitch binds to its cognate ligand with high affinity (~4 nM) and discriminates strongly against adenine (~10,000-fold). The three-dimensional structures of the riboswitch in complex with various ligands showed almost complete encapsulation of ligands accompanied by local conformational changes around the binding pocket and extensive interactions through multiple hydrogen bonds with U22, U47, U51 and C74 (Figure 5a) [36][96][97][37,97,98]. The high specificity to guanine is achieved by Watson–Crick base pairing with the nucleobase C74. Based on these observations, efforts to discover the guanine riboswitch targeting compounds have remained focused on purine analogues.
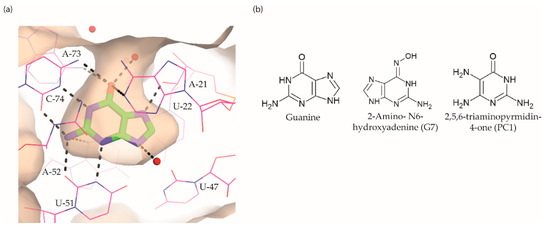
Figure 5.
a
b
Breaker and colleagues were the first to examine the antibacterial activity of guanine analogues [95][96]. Although the study identified 2-amino-N6-hydroxyadenine (Figure 5b) to exert antibacterial activity by targeting the guanine riboswitch, the possibility of this known mutagen hitting off-target(s) could not be excluded [98][99][99,100]. Lafontaine et al. explored the guanine riboswitch by screening various pyrimidine derivatives and guanine analogues modified at the two-, and six-positions or in the five-membered ring [100][101][101,102]. The most promising hit was 2,5,6-triaminopyrimidin-4-one (PC1, Figure 5b). Interestingly, PC1 inhibited the growth of only those clinical strains wherein guanosine monophosphate (GMP) synthetase (guaA) is under riboswitch control, namely S. aureus, S. haemolyticus, methicilin resistant S. aureus COL and C. difficile. Further, in mice, as well as a bovine model of S. aureus infection, PC1 reduced the bacterial burden in a dose-dependent manner [100][102][101,103]. However, it is unclear if the antibacterial effect is only due to binding to the guanine riboswitch. One concern is that no resistant bacteria were obtained even after > 30 passages of a S. aureus strain containing functional guaA regulated by a riboswitch [100][101], which can mean that PC1 has multiple targets. Further, in a different study it was found that guanine and GMP only marginally rescued bacterial growth in the presence of PC1, implying that the guanine pathway is not the sole target of this compound [103][104]. It should, however, be noted that different S. aureus strains were used in these studies so that strain-dependent variations in response to PC1 cannot be excluded. However, PC1 was also shown to be cytotoxic to macrophages, which lack a guanine riboswitch further suggesting that PC1 has other targets besides the guanine riboswitch. Regardless, the discovery of PC1 advocates the possibility of targeting the guanine riboswitch with compounds containing a distinct chemical scaffold from the cognate ligand. As the guanine riboswitch regulates genes involved in de novo guanine biosynthesis—guaA and guaB—which are only present in a few clinical pathogenic strains, the riboswitch is considered a target for narrow-spectrum antibacterials.
