With many successful stories, machine learning (ML) and deep learning (DL) have been widely used in our everyday lives in a number of ways. They have also been instrumental in tackling the outbreak of Coronavirus (COVID-19), which has been happening around the world. The SARS-CoV-2 virus-induced COVID-19 epidemic has spread rapidly across the world, leading to international outbreaks. The COVID-19 fight to curb the spread of the disease involves most states, companies, and scientific research institutions. In this research, we look at the Artificial Intelligence (AI)-based ML and DL methods for COVID-19 diagnosis and treatment. Furthermore, in the battle against COVID-19, we summarize the AI-based ML and DL methods and the available datasets, tools, and performance.
- COVID-19
- diagnosis
- treatment
- artificial intelligence
- machine learning
- deep learning
1. Introduction
Severe Acute Respiratory Syndrome Corona-Virus 2 (SARS-CoV-2) is currently a considerable infectious worldwide disease. Corona-virus 2019 (COVID-19), which was caused by a virus named SARS-CoV-2, was very first apprised in Wuhan, China in December 2019 and later in many parts all over the world; on 3 January 2020, the World Health Organization declared that COVID-19 is a Public Health Emergency of International Concern (PHEIC), and confirmed it as an epidemic on 11 March 2020 [1]. This disease has been accounted for in 216 countries and regions all around from 16 May 2020. The disease has spread and prompted momentous side effects, with 86,159,886 cases of confirmed coronavirus and 1,861,764 deaths on 5 January 2021.
The health industry is eagerly looking for new technologies and techniques to track and control the growth of coronavirus epidemic in this international health crisis. One of the greatest uses global technology right now is Artificial Intelligence (AI), which can track the speed and detect the growth rate of the corona virus, and identify the risk and severity of Corona virus patients. AI can also anticipate the possibility of death by adequately analysing previous patient data. Artificial intelligence can assist us in battling the virus by testing individuals, medical assistance, data and information, and recommendations regarding disease control.
In order to solve complex problems in our lives, AI is a broad umbrella that consists of many sub-areas. These sub-areas include learning, preparation, thinking, representation of information, and searching. Machine Learning (ML) and Deep Learning (DL) are a subset of AI areas that consist of several algorithms that provide intelligent models to identify or cluster particular tasks.
ML is a subset of AI that consists in the algorithmic modeling culture of statistical models [2], and only needs a small amount of knowledge to learn how to solve problems. Logistic Regression (LR), Decision Tree (DT), Random Forest (RF), K-nearest Neighbor (KNN), Adaboost, K-means clustering (KC), Density clustering (DC), Hidden Markov Models (HMM), Support vector machine (SVM), Naive Bayes (NB), Restricted Boltzmann Machines (RBM), and Artificial Neural Network (ANN), such as Recurrent Neural Networks (RNN), including Long-short-term-memory (LSTM), Autoencoder (AE), and Generative Adversarial Network (GAN), are ML techniques.
DL, on the other hand, is a subset of ML that focuses on building deep structural NN models that learn from data using algorithms of feedforward and backprobagation. After ML, the DL emerged and outperformed it in the last two decades in several activities. Nevertheless, it takes a huge amount of data to understand. Exceptional cases of DL, where large-scale data are not needed to train, have been transfer learning and generative models. DL algorithms usually involve Deep Belief Networks (DBN), Deep Neural Network (DNN), and Deep Convolutional Neural Networks (Deep CNN).
Appreciatively, research works in industry, medical, technological, and military sectors have victoriously introduced advanced AI-based ML and DL methods in the COVID-19 war within a short period after the outbreak of COVID-19 and attained substantial progress. For example, across medical image analysis, ML and DL help COVID-19 diagnosis as well as provide non-invasive detection measures to avoid medical personnel from contracting pathogens and, for further treatment, the patient’s severity score is also given. In virology studies, ML and DL are used to examine SARS-CoV-2 protein-related genetics and predict novel combinations that can be used for drug production and vaccination. In addition, on a large-scale, COVID-19 case data and social media data, AI intelligent models that are based on ML and DL learn to construct disease transmission models that accurately predict outbreaks, transmission path, transmission list, and effects. ML and DL are also vastly used in epidemic protection and public monitoring, such as security check-ups in airports, patient tracking, and epidemic detection.
2. Medical Image Inception Using AI-Based ML and DL for the Detection of COVID-19
2.2. Chest X-ray Image Detection
Chest X-rays have been proposed as a highly helpful method for evaluating and testing COVID-19 patients.
Figure 2 shows representative architectures of DL-based CT image classification and COVID-19 examination. When compared with CT images, chest X-ray (CXR) images are simpler to acquire in clinical radiology examinations. There are many available studies [14,15] that operates on chest X-ray (CXR) images for corona virus detection. For the most part, the CXR image testing factor that is based on AI strategies involves measures, such as data correction, model training, and segmentation of COVID-19. There are several methodologies of deep learning (such as CNN, nCOVnet, and U-Net++) that are used to find better and fast detection in the detection of COVID-19 on X-ray images.
shows representative architectures of DL-based CT image classification and COVID-19 examination. When compared with CT images, chest X-ray (CXR) images are simpler to acquire in clinical radiology examinations. There are many available studies [3][4] that operates on chest X-ray (CXR) images for corona virus detection. For the most part, the CXR image testing factor that is based on AI strategies involves measures, such as data correction, model training, and segmentation of COVID-19. There are several methodologies of deep learning (such as CNN, nCOVnet, and U-Net++) that are used to find better and fast detection in the detection of COVID-19 on X-ray images.
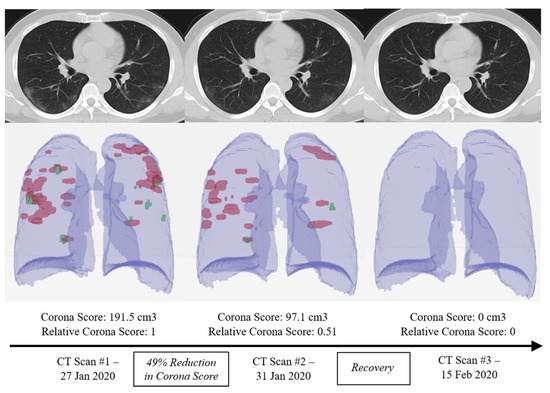
Figure 2. Using different computed tomography (CT) scans from a a COVID-19 patient, the RADLogics algorithm measures the recovery amount using a specific score [16][5].
2.3. COVID-19 Severity Classification Using Chest X-ray with Deep Learning
With the support of artificial intelligence, chest X-rays enable us to understand more, particularly by using ML and DL techniques. Chest X-rays (CXRs) offer a non-invasive method for monitoring disease progression. For front-chest X-ray pictures, a severity score predictor model of COVID-19 pneumonia is being studied in [48]. Expansions of lung involvement and light intensity are also included in the CXR image database. A pre-trained neural network model (i.e., DenseNet) in large chest X-ray sets (non-COVID-19) is used in order to create features of COVID-19 images to predict this activity. 94 images of COVID-19 certified patients go to studying the severity of COVID-19 prediction while using DL, as shown in
With the support of artificial intelligence, chest X-rays enable us to understand more, particularly by using ML and DL techniques. Chest X-rays (CXRs) offer a non-invasive method for monitoring disease progression. For front-chest X-ray pictures, a severity score predictor model of COVID-19 pneumonia is being studied in [6]. Expansions of lung involvement and light intensity are also included in the CXR image database. A pre-trained neural network model (i.e., DenseNet) in large chest X-ray sets (non-COVID-19) is used in order to create features of COVID-19 images to predict this activity. 94 images of COVID-19 certified patients go to studying the severity of COVID-19 prediction while using DL, as shown in
. A score-based methodology is used, which includes two forms of scores.
provides examples of disease severity stages: the level of lung participation and the degree of ambiguity are shown at a time in a single X-ray image.
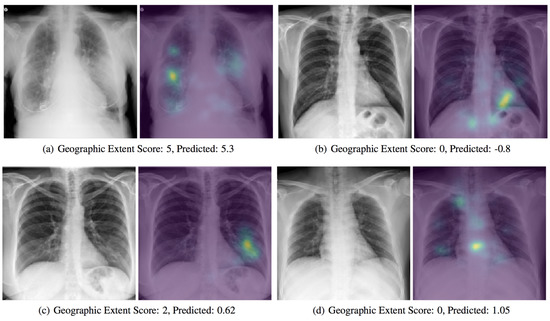
Figure 4. A presentation of a predicted severity scores for COVID-19 chest X-ray scans while using the DenseNet model [48][6].
Table 2.
COVID-19 severity stages using a score-based system.
| Parameters | Severity Score | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| Extent of Lung Involvement | No | <25% | 25–50% | 50–75% | >75% |
| Degree of ambiguity | No | Ground glass ambiguity | Unification | White-out | - |
3. ML and DL for Drug and Vaccine Development
In combination with a large amount of data, the capacity for automatic abstract component learning has had a major impact on the efficacious use of ML. Drug discovery and vaccination affected two fields of high importance, where ML provided integrated property predictions, behaviour prediction, reaction prediction, and ligand-protein interactions. mDiverse drug development programs and vaccines for SARS-CoV-2 and COVID-19 have been suggested to focus on proteomics and genomics studies. One of the main contributions to intelligent medicine is the use of ML and DL in the development of new medicines and vaccines, and it plays a major function in the battle against COVID-19.
3.1. ML and DL For Vaccine Development
In this context, ML and DL play two important supporting roles: the dissemination of vaccine components by observing the viral protein structure, and helping medical researchers to review a large number of important research papers at an incredible rate. Three main types of vaccines are available: vaccines for any pathogen, such as flu or MMR, use deadly or compromised immune system infections; subunit vaccines (e.g., pertussis, shingles) only use part of the virus, such as protein; and, vaccines for nucleic acid inject viral genes into human cells to improve the response of the body [71]. The newest is the COVID-19 vaccine, which began trials in the United States this week. AI helps to speed up the growth of subunits and nucleic acids.
In this context, ML and DL play two important supporting roles: the dissemination of vaccine components by observing the viral protein structure, and helping medical researchers to review a large number of important research papers at an incredible rate. Three main types of vaccines are available: vaccines for any pathogen, such as flu or MMR, use deadly or compromised immune system infections; subunit vaccines (e.g., pertussis, shingles) only use part of the virus, such as protein; and, vaccines for nucleic acid inject viral genes into human cells to improve the response of the body [7]. The newest is the COVID-19 vaccine, which began trials in the United States this week. AI helps to speed up the growth of subunits and nucleic acids.
In analysing how it functions, understanding protein composition is critical. Researchers can produce drugs that work in various protein shapes once the situation is acknowledged. However, testing any protein structure will take quite a long time before finding its unique 3D structure. The method of evaluating protein structure and its genetic sequence can be simplified by AI systems that are based on DL.
Google DeepMind [72,73] introduced AlphaFold in January, an advanced and specialised system that forecasts the formation of 3D proteins while using their genetic sequence. In the beginning of March, the system was tested in COVID-19. In order to asses the scientific community for understanding the virus, DeepMind has published a protein prediction for several untreated proteins that are related to SARS-CoV-2, since it is the main cause for COVID-19. Meanwhile, scientists at the University of Texas at Austin and the National Institutes of Health have applied a standard biological method to establish the first 3D atomic map on the scale of a spike protein component of a virus that binds to human cells. Other Coronaviruses, including SARS-CoV and MERS-CoV, have spent years collaborating with the team that is responsible for this crucial breakthrough. Accurate predictions for this spike structure were provided by one of the predictions released by AlphaFold. Computer simulations for building 3D atom models on the SARS-CoV-2 protein scale neatly related to the results that were found on the UT Austin board were also used by [74] at the Institute for Protein Design at the University of Washington. By constructing new proteins to minimize Coronavirus, they are currently continuing on this work. In principle, these proteins will conform to a protein spike that protects healthy cells from being invaded by viral particles.
Google DeepMind [8][9] introduced AlphaFold in January, an advanced and specialised system that forecasts the formation of 3D proteins while using their genetic sequence. In the beginning of March, the system was tested in COVID-19. In order to asses the scientific community for understanding the virus, DeepMind has published a protein prediction for several untreated proteins that are related to SARS-CoV-2, since it is the main cause for COVID-19. Meanwhile, scientists at the University of Texas at Austin and the National Institutes of Health have applied a standard biological method to establish the first 3D atomic map on the scale of a spike protein component of a virus that binds to human cells. Other Coronaviruses, including SARS-CoV and MERS-CoV, have spent years collaborating with the team that is responsible for this crucial breakthrough. Accurate predictions for this spike structure were provided by one of the predictions released by AlphaFold. Computer simulations for building 3D atom models on the SARS-CoV-2 protein scale neatly related to the results that were found on the UT Austin board were also used by [10] at the Institute for Protein Design at the University of Washington. By constructing new proteins to minimize Coronavirus, they are currently continuing on this work. In principle, these proteins will conform to a protein spike that protects healthy cells from being invaded by viral particles.
Researchers merged AI with cloud computing to stop the Spike protein from binding to the ACE2 receptor in human cells and to produce a possible vaccine for COVID-19 [75]. Flinders University researchers studied the COVID-19 virus and then applied their data to model a vaccine, called Covax-19, as shown in
Researchers merged AI with cloud computing to stop the Spike protein from binding to the ACE2 receptor in human cells and to produce a possible vaccine for COVID-19 [11]. Flinders University researchers studied the COVID-19 virus and then applied their data to model a vaccine, called Covax-19, as shown in
.
Figure 6. Covax-19™ is an Australian-designed COVID-19 vaccine modeled by AI-based technologies [76].
Covax-19™ is an Australian-designed COVID-19 vaccine modeled by AI-based technologies [12].
In order to determine how the virus was harming human cells, the researchers utilized computer generated models of the S protein and its human receptor, the enzyme-converting angiotensin 2 (ACE2). Subsequently, they attempted to produce a vaccine that could prevent this mechanism. The team used the most innovative AI and advanced cloud computing technology to speed up the production of vaccines [73,75,77].
In order to determine how the virus was harming human cells, the researchers utilized computer generated models of the S protein and its human receptor, the enzyme-converting angiotensin 2 (ACE2). Subsequently, they attempted to produce a vaccine that could prevent this mechanism. The team used the most innovative AI and advanced cloud computing technology to speed up the production of vaccines [9][11][13].
3.2. Drug Development
With the development of a predictive learning model, AI systems that are based on ML and DL are used in field design and they perform a fast-paced test to accurately represent the performance. The AI systems can easily classify drugs that can battle infectious diseases, such as COVID-19, with a drug recovery process. With this breakthrough, which is a medical evidence-based tool, will potentially enhance drug accessibility, preparation, care, and recorded patient outcomes COVID-19. Drug development is a very dangerous, lengthy, and costly phase. While it takes ten to fifteen years to make a new molecular venture, the success rate is only 2.01%, as reported by the Eastern Research Group (ERG) [84]. For the treatment of a never-considered medicinal indication, the idea of drug repurposing reuses old medications. It is an experimental strategy for the detection of pre-approved, discontinued, shelved, and investigational drugs for the treatment of other diseases with approved restatement. The development of conventional drugs generally involves five steps [85]:
With the development of a predictive learning model, AI systems that are based on ML and DL are used in field design and they perform a fast-paced test to accurately represent the performance. The AI systems can easily classify drugs that can battle infectious diseases, such as COVID-19, with a drug recovery process. With this breakthrough, which is a medical evidence-based tool, will potentially enhance drug accessibility, preparation, care, and recorded patient outcomes COVID-19. Drug development is a very dangerous, lengthy, and costly phase. While it takes ten to fifteen years to make a new molecular venture, the success rate is only 2.01%, as reported by the Eastern Research Group (ERG) [14]. For the treatment of a never-considered medicinal indication, the idea of drug repurposing reuses old medications. It is an experimental strategy for the detection of pre-approved, discontinued, shelved, and investigational drugs for the treatment of other diseases with approved restatement. The development of conventional drugs generally involves five steps [15]:
-
Drug discovery and development.
Drug pre-clinical research.
Drug clinical research.
FDA drug review.
FDA drug after-market safety control and development.
But only drug repurposing requires four steps [85]:
But only drug repurposing requires four steps [15]:
-
Compound identification.
Compound acquisition.
Drug clinical research.
FDA drug after-market safety control and development.
The discovery and development of new molecular structure are slow, time consuming, and expensive. Therefore, the best option is to reconstruct the approved drugs for SARS-COV-2 therapy. In this case, in the treatment of viral infection, Chloroquine (CQ) and its hydroxyl analogue Hydroxychloroquine (HCQ) have been identified. These drugs have antimalarial efficacy and they have also been shown to treat COVID-19 in vitro [86].
The discovery and development of new molecular structure are slow, time consuming, and expensive. Therefore, the best option is to reconstruct the approved drugs for SARS-COV-2 therapy. In this case, in the treatment of viral infection, Chloroquine (CQ) and its hydroxyl analogue Hydroxychloroquine (HCQ) have been identified. These drugs have antimalarial efficacy and they have also been shown to treat COVID-19 in vitro [16].
One AI-based drug development team focuses on the discovery at the molecular level of new drug-like compounds. Beck et al. [87] suggested a DL-based drug-target interaction model, called Molecule Transformer-Drug Target Interaction (MT-DTI), to predict potential drugs for COVID-19. SMILES strings and amino-acid sequences were used in the MT-DTI model in order to classify target proteins with 3D crystal structures. The findings showed that atazanavir, an antiretroviral drug that is used to treat and prevent the human immunodeficiency virus (HIV), is the best chemical compound with an inhibitory potency with Kd of 94.94 nM against 3C-like SARS-CoV-2 proteinase, followed by remdesivir (113.13 nM), efavirenz (199.17 nM), ritonavir (204.05 nM), and dolutegravir (336.91 nM). From the databases of NCBI, Drug Target Popular (DTC), and BindingDB, the authors obtained the amino acid sequences of 3C-like proteases and associated antiviral drugs and drug targets. In addition, they utilised a molecular docking and virtual screening method (AutoDock Vina) in order to determine the binding affinity between 3410 drugs and SARS-CoV-2 3CLpro. Six possible medicines, such as Remdesivir, Atazanavir, Efavirenz, Ritonavir, Dolutegravir, Kaletra (lopinavir/ritonavir), produced experimental results. Note that, in a clinical trial, Remdesivir looks promising.
One AI-based drug development team focuses on the discovery at the molecular level of new drug-like compounds. Beck et al. [17] suggested a DL-based drug-target interaction model, called Molecule Transformer-Drug Target Interaction (MT-DTI), to predict potential drugs for COVID-19. SMILES strings and amino-acid sequences were used in the MT-DTI model in order to classify target proteins with 3D crystal structures. The findings showed that atazanavir, an antiretroviral drug that is used to treat and prevent the human immunodeficiency virus (HIV), is the best chemical compound with an inhibitory potency with Kd of 94.94 nM against 3C-like SARS-CoV-2 proteinase, followed by remdesivir (113.13 nM), efavirenz (199.17 nM), ritonavir (204.05 nM), and dolutegravir (336.91 nM). From the databases of NCBI, Drug Target Popular (DTC), and BindingDB, the authors obtained the amino acid sequences of 3C-like proteases and associated antiviral drugs and drug targets. In addition, they utilised a molecular docking and virtual screening method (AutoDock Vina) in order to determine the binding affinity between 3410 drugs and SARS-CoV-2 3CLpro. Six possible medicines, such as Remdesivir, Atazanavir, Efavirenz, Ritonavir, Dolutegravir, Kaletra (lopinavir/ritonavir), produced experimental results. Note that, in a clinical trial, Remdesivir looks promising.
Using effective machine learning techniques, two similarity-based methods, KronRLS [88] and SimBoost [89], have been proposed. However, there are two downsides to this matrix. First, the representation of features is reduced, which would cause the prediction faulty. Second, it involves the estimation of the matrix of similarity, which, in the training phase, will limit the maximum number of molecules. A deep learning based DTI model, DeepDTA [90], was proposed in order to address these limitations. It is a CNN-based end-to-end model that waives the feature engineering requirement. From a raw molecule and protein sequence, the model automatically finds useful features. The performance was seen on two publicly accessible DTI benchmarks, i.e., Sim-Boost and KronRLS. In order to extract features representations from the raw protein sequences and SMILES strings and train them, they used CNN blocks and combined these features representations to input into a deep CNN, naming it DeepDTA. They applied Smith–Waterman (S-W) and Pubchem Similarity algorithms to process the proteins and ligands’ pair-wise similarities, respectively. In tarining the patterns of the data, three alternative combinations applied this knowledge as input to the proposed ad enhanced DeepDTA model. The following are the three alternative combinations for training this model:
Using effective machine learning techniques, two similarity-based methods, KronRLS [18] and SimBoost [19], have been proposed. However, there are two downsides to this matrix. First, the representation of features is reduced, which would cause the prediction faulty. Second, it involves the estimation of the matrix of similarity, which, in the training phase, will limit the maximum number of molecules. A deep learning based DTI model, DeepDTA [20], was proposed in order to address these limitations. It is a CNN-based end-to-end model that waives the feature engineering requirement. From a raw molecule and protein sequence, the model automatically finds useful features. The performance was seen on two publicly accessible DTI benchmarks, i.e., Sim-Boost and KronRLS. In order to extract features representations from the raw protein sequences and SMILES strings and train them, they used CNN blocks and combined these features representations to input into a deep CNN, naming it DeepDTA. They applied Smith–Waterman (S-W) and Pubchem Similarity algorithms to process the proteins and ligands’ pair-wise similarities, respectively. In tarining the patterns of the data, three alternative combinations applied this knowledge as input to the proposed ad enhanced DeepDTA model. The following are the three alternative combinations for training this model:
Training only compound representation.
Training only protein sequence representation.
-
Training both protein representation and compound representations.
References
- Soghaier, M.A.; Saeed, K.M.; Zaman, K.K. Public Health Emergency of International Concern (PHEIC) has declared twice in 2014; polio and Ebola at the top. AIMS Public Health 2015, 2, 218.
- Breiman, L. Statistical modeling: The two cultures (with comments and a rejoinder by the author). Stat. Sci. 2001, 16, 199–231.
- Pandit, M.K.; Banday, S.A. SARS n-CoV2-19 detection from chest X-ray images using deep neural networks. Int. J. Pervasive Comput. Commun. 2020, 16, 419–427.
- Basu, S.; Mitra, S. Deep Learning for Screening COVID-19 using Chest X-ray Images. arXiv 2020, arXiv:2004.10507.
- Scudellari, M. Hospitals Deploy AI Tools to Detect COVID-19 on Chest Scans. 2020. Available online: https://spectrum.ieee.org/the-human-os/biomedical/imaging/hospitals-deploy-ai-tools-detect-covid19-chest-scans (accessed on 10 September 2020).
- Cohen, J.P.; Dao, L.; Morrison, P.; Roth, K.; Bengio, Y.; Shen, B.; Abbasi, A.; Hoshmand-Kochi, M.; Ghassemi, M.; Li, H.; et al. Predicting covid-19 pneumonia severity on chest X-ray with deep learning. arXiv 2020, arXiv:2005.11856.
- Etzioni, O.; Decario, N. AI Can Help Scientists Find a Covid-19 Vaccine. 2020. Available online: https://www.wired.com/story/opinion-ai-can-help-find-scientists-find-a-covid-19-vaccine/ (accessed on 16 September 2020).
- Senior, A.; Jumper, J.; Hassabis, D.; Kohli, P. AlphaFold: Using AI for Scientific Discovery. DeepMind. 2018. Available online: https://deepmind.com/blog/alphafold (accessed on 28 October 2020).
- HospiMedica. Scientists Use Cloud-Based Supercomputing and AI to Develop COVID-19 Treatments and Vaccine Models. 2020. Available online: https://www.hospimedica.com/covid-19/articles/294784537/scientists-use-cloud-based-supercomputing-and-ai-to-develop-covid-19-treatments-and-vaccine-models.html (accessed on 15 September 2020).
- Institute for Protein Design. Rosetta’s Role in Fighting Coronavirus. 2020. Available online: https://www.ipd.uw.edu/2020/02/rosettas-role-in-fighting-coronavirus/ (accessed on 10 September 2020).
- Rees, V. AI and Cloud Computing Used to Develop COVID-19 Vaccine. 2020. Available online: https://www.drugtargetreview.com/news/59650/ai-and-cloud-computing-used-to-develop-covid-19-vaccine/ (accessed on 17 September 2020).
- TABIP. Coronavirus Vaccine Candidate Developed In Adelaide Lab To Start Human Trials. 2020. Available online: https://covid19.tabipacademy.com/2020/07/03/coronavirus-vaccine-candidate-developed-in-adelaide-lab-to-start-human-trials/ (accessed on 20 September 2020).
- Flinders University. Microsoft’s AI for Health Supports COVID-19 Vaccine. 2020. Available online: https://medicalxpress.com/news/2020-08-microsoft-ai-health-covid-vaccine.html (accessed on 18 September 2020).
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of drug repositioning approaches and resources. Int. J. Biol. Sci. 2018, 14, 1232.
- Mohanty, S.; Rashid, M.H.A.; Mridul, M.; Mohanty, C.; Swayamsiddha, S. Application of Artificial Intelligence in COVID-19 drug repurposing. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1027–1031.
- Khuroo, M.S.; Sofi, A.A.; Khuroo, M. Chloroquine and Hydroxychloroquine in Coronavirus Disease 2019 (COVID-19). Facts, Fiction & the Hype. A Critical Appraisal. Int. J. Antimicrob. Agents 2020, 56, 106101.
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790.
- Pahikkala, T.; Airola, A.; Pietilä, S.; Shakyawar, S.; Szwajda, A.; Tang, J.; Aittokallio, T. Toward more realistic drug–target interaction predictions. Brief. Bioinform. 2015, 16, 325–337.
- He, T.; Heidemeyer, M.; Ban, F.; Cherkasov, A.; Ester, M. SimBoost: A read-across approach for predicting drug–target binding affinities using gradient boosting machines. J. Cheminform. 2017, 9, 1–14.
- Öztürk, H.; Özgür, A.; Ozkirimli, E. DeepDTA: Deep drug–target binding affinity prediction. Bioinformatics 2018, 34, i821–i829.
