ADP-ribosyltransferases (ARTs) are a well-known type of bacterial toxin [2]. They transfer an ADP-ribose moiety from nicotinamide adenine dinucleotide (NAD+) onto a target protein to generate an ADP-ribosylated protein and nicotinamide. The bulky and negatively charged ADP-ribose moiety affects the protein function by sterically blocking interactions with partner molecules, inducing conformational changes, or creating docking sites for new interaction partners [3].
- ADP-ribosyltransferase
- substrate recognition
- bacterial toxin
1. Introduction
ARTs have been implicated in a wide range of processes in organisms from all kingdoms of life, including the toxin–antitoxin system of bacteria and the DNA damage repair, transcription, cell-cycle progression, and cell division processes of eukaryotes [4][1]. Individual ARTs have specific substrates and modify specific residues or functional groups of not only proteins but also DNA, RNA, and antibiotics [5][2]. ARTs are subdivided into two classes based on their conservation of three significant motifs: the R-S-E class, which is related to cholera toxin; and the H-Y-E class, which is related to diphtheria toxin (Figure 1a) [6][3].
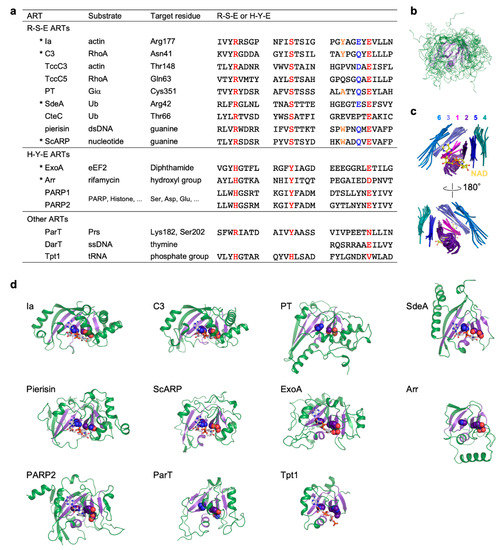
Figure 1. Comparison of R-S-E class, H-Y-E class, and other ADP-ribosyltransferases. (a) Substrate, target residue, and conserved motifs. Conserved R-S-E and H-Y-E residues are shown in red. In the R-S-E motif, the third aromatic residue and the sixth glutamate or glutamine residue in the ADP-ribosylating toxin turn-turn (ARTT) loop (X-X-ϕ-X-X-E/Q-X-E) are shown in orange and blue, respectively. ADP-ribosyltransferases (ARTs) with available substrate-complexed structures are indicated with asterisks. (b) Superimposed structures of the ART domains of the 11 ARTs are shown in (d). The structure of the ART core, including six strands and the helix, are shown in violet. (c) Close-up views of ART core structures. Individual strands are shown in different colors and are numbered from the N-terminus to the C-terminus. NAD bound to Ia is shown as a ball-and-stick model. (d) Structures of the ART domain. ART core structures are shown in violet. R-S-E and H-Y-E residues are shown as sphere models. NAD or NAD analogs are shown as stick models. Protein Data Bank (PDB) IDs: Ia, 4h03; C3, 4xsh; PT, 1bcp; SdeA, 5yij; Pierisin, 5h6j; ScARP, 5zj5; ExoA, 2zit; Arr, 2hw2; PARP2, 6tx3; ParT, 6d0h; Tpt1, 6e3a.
ARTs transfer an ADP-ribose moiety specifically onto nucleophilic atoms as follows: onto the oxygen atom of the carboxyl groups of aspartate and glutamate, the hydroxyl groups of serine and antibiotics, and the 5′ phosphate group of double-stranded DNA (dsDNA); onto the nitrogen atom of the amino group of the guanidino groups of arginine and guanine, the imidazole group of diphthamide, and the carboxamide groups of asparagine and glutamine; and onto the sulfur atom of the thiol group of cysteine [7][4]. However, the precise mechanism by which ARTs modify a specific residue or functional group remains an open question.
2. ART Structures
Most ARTs can be subdivided into the R-S-E and H-Y-E classes, but some do not belong to either class (Figure 1a). However, superimposition of ART domains of all the ARTs shows that there is strict conservation of the core structure consisting of six strands and a helix (Figure 1b–d) [6][3]. The R-S-E and H-Y-E residues are located on strands of the core structure: The arginine and histidine, serine and tyrosine, and glutamate residues are located on the first, second, and fifth strands from the N-terminus, respectively. NAD binds to the cleft formed by the core structure. The arginine and histidine residues of the R-S-E and H-Y-E motifs are important for NAD binding, while the glutamate residues are essential for the cleavage of NAD+.
3. Substrates and Target Residues of R-S-E Class ARTs
Most ARTs that act as bacterial toxins belong to the R-S-E class, and many members of this class have been found to affect the actin cytoskeleton. Iota toxin (Ia) from Clostridium perfringens specifically modifies actin on Arg177; although the modified actin structure does not show any major conformational change compared to non-modified actin, the modified actin is not able to polymerize due to the steric hindrance from the ADP-ribose moiety, resulting in actin depolymerization and cell rounding [8][5]. Many Ia-homologous toxins are known: C2I from Clostridium botulinum [9][6], CDTa from Clostridium difficile [10][7], CSTa from Clostridium spiroforme [11][8], VIP2 from Bacillus cereus [12][9], and CPILEa (also known as BECa) from C. perfringens [13,14][10][11]. All of these toxins ADP-ribosylate Arg177 on actin [15] [12]. C3 toxin from Clostridium botulinum specifically modifies Rho GTPase, the master regulator of the actin cytoskeleton, on Asn41 [16][13]. Rho GTPase functions as a molecular switch between active GTP-bound Rho, which transduces signals by interacting with effector proteins and inactive GDP-bound Rho. ADP-ribosylation occurs on both GTP- and GDP-bound Rho. Although the modified GTP-bound Rho is still able to bind its effector proteins [17][14], the modified GDP-bound Rho shows a tighter binding with guanine nucleotide dissociation inhibitor (GDI) compared to non-modified Rho, which prevents activation and results in the destruction of actin stress fibers [18][15]. Many C3-homologous toxins have been described, including C3lim from Clostridium limosum [19][16], C3cer from Bacillus cereus [20][17], C3stau from Staphylococcus aureus [21][18], and C3larvinA and Plx2A from Paenibacillus larvae [22,23][19][20]. The components of the Tc Toxin complex from Photorhabdus luminescens, TccC3 and TccC5, specifically modify actin on Thr148 (a different target residue than that of iota toxin) and RhoA on Gln63 (a different target residue than that of C3 toxin), respectively [24][21]. Because the modified residues are different, the functional consequences of TccC3 and TccC5 are different from those of iota and C3: TccC3 and TccC5 do not cause actin depolymerization but rather its polymerization or clustering. Thr148 of actin is located in the site of its interaction with thymosin-β4, which prevents actin polymerization. ADP-ribosylation of actin at Thr148 reduces the affinity for thymosin-β4 to inhibit actin sequestration and promote actin polymerization. Gln63 of RhoA is essential for the hydrolysis of GTP; the ADP-ribosylation of RhoA at Gln63 causes its persistent activation, resulting in strong stress fiber formation. Studies have also shown that ARTs are utilized as toxins for interbacterial competition [25][22]. Tre1 from Serratia proteamaculans, which is an effector protein via the type VI secretion system, specifically ADP-ribosylates Arg174 on the bacterial tubulin-like protein FtsZ. This ADP-ribosylation disrupts cell division, leading to cell elongation and cell death. Because the effector is delivered indiscriminately, S. proteamaculans has a means to prevent self-intoxication: The cognate immunity protein, Tri1, protects against Tre1 toxicity by physical sequestration and enzymatic ADP-ribose cleavage.
Cysteine-targeting ADP-ribosylation is unusual in ARTs, but pertussis toxin from Bordetella pertussis specifically modifies Cys351 on the α subunit of inhibitory trimeric G-protein (Giα), which is a conserved cysteine located in the fourth position from the C-terminus [26][23]. Because this ADP-ribosylation prevents the binding to G protein-coupled receptor (GPCR) due to steric hindrance, Giα remains in its inactive GDP-bound state and is unable to inhibit adenylate cyclase activity, resulting in elevation of the intracellular cAMP level. Pertussis-like toxin from Escherichia coli (EcPlt) has been also analyzed: Rather than targeting Cys351, this toxin was found to target Lys345 and Asn347 [27][24].
Ubiquitin-targeting R-S-E class ARTs have been also discovered. The SidE family effector proteins from Legionella pneumophila—SdeA, SdeB, SdeC, and SidE—specifically modify ubiquitin (Ub) on Arg42 [28,29][25][26]. SdeA consists of N-terminal deubiquitinase (DUB), phosphodiesterase (PDE), ART, and coiled-coil (CC) domains. SdeA catalyzes ADP-ribosylation of Ub on Arg42 via its ART domain and then cleaves the pyrophosphate bond of the ADP-ribose moiety via its PDE domain to release AMP. The cleavage is coupled to a nucleophilic attack of water or a serine residue of a substrate protein, which generates phosphoribosylated Ub or a ubiquitinated protein via phosphoribose, respectively. This ubiquitination of a substrate protein is totally different from the conventional ubiquitination that proceeds through the conserved enzymes E1, E2, and E3. ADP-ribosylation and phosphoribosylation of ubiquitin prevent the reaction with E1 due to steric hindrance, which impairs several Ub-dependent processes. The type III effector, CteC, from Chromobacterium violaceum specifically ADP-ribosylates Thr66 of ubiquitin, which prevents the transfer of ubiquitin from E1 to E2 and thereby disrupts ubiquitin signaling [30][27]. CteC modifies both mono- and poly-Ub. The ADP-ribosylation of mono-Ub does not affect E1-mediated Ub activation but prevents the transfer of Ub from E1 to E2, leading to inhibition of poly-Ub chain synthesis. The ADP-ribosylation of poly-Ub prevents recognition by Ub binding domains and inhibits deubiquitinase-mediated deubiquitination, leading to dysfunction of poly-Ub chains. Two homologous proteins have been found to show the same function as CteC: CHBU from Burkholderia ubonensis and CHCS from Corallococcus show 66% and 24% sequence identity with CteC, respectively. The amino acid sequences of the CteC family members are not similar to those of other R-S-E class ARTs, and their tertiary structure has not yet been revealed.
Pierisin, which was originally purified from larvae of cabbage butterfly Pieris rapae, specifically modify dsDNA on the N2 amino group of the guanine base, causing mutations and an apoptotic response in cancer cells such as the HeLa and TMK-1 cell lines [31,32][28][29]. Pierisin may induce programmed cell death in larval cells to drive insect development, as it is found at a high level (0.4% of total protein) in the pupae of P. rapae [33][30]. It has also been speculated that pierisin functions as a defense factor against parasitization by wasps. ScARP from Streptomyces coelicolor can ADP-ribosylate the N2 amino group of the guanine bases of nucleosides and nucleotides, such as guanosine, deoxyguanosine, GTP, and deoxyGTP, whereas it cannot act against dsDNA [34][31]. Scabin from Streptomyces scabies, which is a putative virulence factor for plants such as potato, also ADP-ribosylates the N2 amino group of the guanine bases of nucleosides, nucleotides, single-stranded DNA, and genomic DNA [35][32]. Mosquitocidal toxin (MTX) from Bacillus sphaericus, which has 30% sequence identity with pierisin, does not target DNA but rather elongation factor Tu (EF-Tu) in Escherichia coli. [36].[33] ADP-ribosylation of EF-Tu prevents the ternary complex formation of EF-Tu, GTP, and aminoacyl-tRNA, resulting in inhibition of bacterial protein synthesis.
4. Substrates and Target Residues of H-Y-E Class ARTs
Exotoxin A (ExoA) from Pseudomonas aeruginosa, diphtheria toxin (DT) from Corynebacterium diphtheriae, and cholix toxin from Vibrio cholera specifically modify translation elongation factor (eEF2) on diphthamide699 in yeast or diphthamide715 in mammals [37,38,39][34][35][36]. ADP-ribosylated eEF2 is unable to mediate the translocation of peptidyl-tRNA from the A to P sites on the ribosome, inhibiting protein synthesis. Arr ADP-ribosylates the hydroxyl group of rifamycins, leading to resistance against these antibiotics [40][37]. Arr is widely distributed in the genomes of pathogenic and nonpathogenic bacteria. Although Arr does not show sequence similarity to known ARTs, its structure is similar to those of known ARTs. The glutamate residue in the conserved motif H-Y-E is replaced by an aspartate residue that is located on the fourth strand (rather than the fifth strand) from the N-terminus. The poly(ADP-ribose) polymerase (PARP) family comprises 17 members: PARP1 to PARP16, which includes two tankyrases (PARP5a and PARP5b) [41,42][38][39]. Poly-ADP-ribose is removed by poly(ADP-ribose) glycohydrolase (PARG) and macrodomain proteins [43][40]. PARPs affect various cellular processes, such as DNA repair, DNA replication, transcriptional regulation, and cell division. The best-studied PARP, PARP1, is thought to be a key player in these processes and has shown promise as a target for anti-cancer drugs.
References
- Palazzo, L.; Mikoč, A.; Ahel, I. ADP-Ribosylation: New Facets of an Ancient Modification. FEBS J. 2017, 284, 2932–2946, doi:10.1111/febs.14078.
- Aravind, L.; Zhang, D.; de Souza, R.F.; Anand, S.; Iyer, L.M. The Natural History of ADP-Ribosyltransferases and the ADP-Ribosylation System. Curr. Top. Microbiol. Immunol. 2015, 384, 3–32, doi:10.1007/82_2014_414.
- Hottiger, M.O.; Hassa, P.O.; Lüscher, B.; Schüler, H.; Koch-Nolte, F. Toward a Unified Nomenclature for Mammalian ADP-Ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219, doi:10.1016/j.tibs.2009.12.003.
- Sung, V.M.-H. Mechanistic Overview of ADP-Ribosylation Reactions. Biochimie 2015, 113, 35–46, doi:10.1016/j.biochi.2015.03.016.
- Margarit, S.M.; Davidson, W.; Frego, L.; Stebbins, C.E. A Steric Antagonism of Actin Polymerization by a Salmonella Viru-lence Protein. Struct. Lond. Engl. 1993 2006, 14, 1219–1229, doi:10.1016/j.str.2006.05.022.
- Aktories, K.; Bärmann, M.; Ohishi, I.; Tsuyama, S.; Jakobs, K.H.; Habermann, E. Botulinum C2 Toxin ADP-Ribosylates Actin. Nature 1986, 322, 390–392, doi:10.1038/322390a0.
- Perelle, S.; Gibert, M.; Bourlioux, P.; Corthier, G.; Popoff, M.R. Production of a Complete Binary Toxin (Actin-Specific ADP-Ribosyltransferase) by Clostridium Difficile CD196. Infect. Immun. 1997, 65, 1402–1407, doi:10.1128/IAI.65.4.1402-1407.1997.
- Simpson, L.L.; Stiles, B.G.; Zepeda, H.; Wilkins, T.D. Production by Clostridium Spiroforme of an Iotalike Toxin That Pos-sesses Mono(ADP-Ribosyl)Transferase Activity: Identification of a Novel Class of ADP-Ribosyltransferases. Infect. Immun. 1989, 57, 255–261, doi:10.1128/IAI.57.1.255-261.1989.
- Han, S.; Craig, J.A.; Putnam, C.D.; Carozzi, N.B.; Tainer, J.A. Evolution and Mechanism from Structures of an ADP-Ribosylating Toxin and NAD Complex. Nat. Struct. Biol. 1999, 6, 932–936, doi:10.1038/13300.
- Irikura, D.; Monma, C.; Suzuki, Y.; Nakama, A.; Kai, A.; Fukui-Miyazaki, A.; Horiguchi, Y.; Yoshinari, T.; Sugita-Konishi, Y.; Kamata, Y. Identification and Characterization of a New Enterotoxin Produced by Clostridium Perfringens Isolated from Food Poisoning Outbreaks. PLoS ONE 2015, 10, e0138183, doi:10.1371/journal.pone.0138183.
- Yonogi, S.; Matsuda, S.; Kawai, T.; Yoda, T.; Harada, T.; Kumeda, Y.; Gotoh, K.; Hiyoshi, H.; Nakamura, S.; Kodama, T.; et al. BEC, a Novel Enterotoxin of Clostridium Perfringens Found in Human Clinical Isolates from Acute Gastroenteritis Out-breaks. Infect. Immun. 2014, 82, 2390–2399, doi:10.1128/IAI.01759-14.
- Aktories, K.; Lang, A.E.; Schwan, C.; Mannherz, H.G. Actin as Target for Modification by Bacterial Protein Toxins. FEBS J. 2011, 278, 4526–4543, doi:10.1111/j.1742-4658.2011.08113.x.
- Aktories, K.; Braun, U.; Rösener, S.; Just, I.; Hall, A. The Rho Gene Product Expressed in E. Coli Is a Substrate of Botulinum ADP-Ribosyltransferase C3. Biochem. Biophys. Res. Commun. 1989, 158, 209–213, doi:10.1016/s0006-291x(89)80199-8.
- Genth, H.; Schmidt, M.; Gerhard, R.; Aktories, K.; Just, I. Activation of Phospholipase D1 by ADP-Ribosylated RhoA. Bio-chem. Biophys. Res. Commun. 2003, 302, 127–132, doi:10.1016/s0006-291x(03)00112-8.
- Genth, H.; Gerhard, R.; Maeda, A.; Amano, M.; Kaibuchi, K.; Aktories, K.; Just, I. Entrapment of Rho ADP-Ribosylated by Clostridium Botulinum C3 Exoenzyme in the Rho-Guanine Nucleotide Dissociation Inhibitor-1 Complex. J. Biol. Chem. 2003, 278, 28523–28527, doi:10.1074/jbc.M301915200.
- Just, I.; Mohr, C.; Schallehn, G.; Menard, L.; Didsbury, J.R.; Vandekerckhove, J.; van Damme, J.; Aktories, K. Purification and Characterization of an ADP-Ribosyltransferase Produced by Clostridium Limosum. J. Biol. Chem. 1992, 267, 10274–10280.
- Wilde, C.; Vogelsgesang, M.; Aktories, K. Rho-Specific Bacillus Cereus ADP-Ribosyltransferase C3cer Cloning and Charac-terization. Biochemistry 2003, 42, 9694–9702, doi:10.1021/bi034583b.
- Wilde, C.; Chhatwal, G.S.; Schmalzing, G.; Aktories, K.; Just, I. A Novel C3-like ADP-Ribosyltransferase from Staphylococ-cus Aureus Modifying RhoE and Rnd3. J. Biol. Chem. 2001, 276, 9537–9542, doi:10.1074/jbc.M011035200.
- Turner, M.; Tremblay, O.; Heney, K.A.; Lugo, M.R.; Ebeling, J.; Genersch, E.; Merrill, A.R. Characterization of C3larvinA, a Novel RhoA-Targeting ADP-Ribosyltransferase Toxin Produced by the Honey Bee Pathogen, Paenibacillus Larvae. Biosci. Rep. 2020, 40, doi:10.1042/BSR20193405.
- Ebeling, J.; Fünfhaus, A.; Knispel, H.; Krska, D.; Ravulapalli, R.; Heney, K.A.; Lugo, M.R.; Merrill, A.R.; Genersch, E. Charac-terization of the Toxin Plx2A, a RhoA-Targeting ADP-Ribosyltransferase Produced by the Honey Bee Pathogen Paenibacil-lus Larvae. Environ. Microbiol. 2017, 19, 5100–5116, doi:10.1111/1462-2920.13989.
- Lang, A.E.; Schmidt, G.; Schlosser, A.; Hey, T.D.; Larrinua, I.M.; Sheets, J.J.; Mannherz, H.G.; Aktories, K. Photorhabdus Lu-minescens Toxins ADP-Ribosylate Actin and RhoA to Force Actin Clustering. Science 2010, 327, 1139–1142, doi:10.1126/science.1184557.
- Ting, S.-Y.; Bosch, D.E.; Mangiameli, S.M.; Radey, M.C.; Huang, S.; Park, Y.-J.; Kelly, K.A.; Filip, S.K.; Goo, Y.A.; Eng, J.K.; et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell 2018, 175, 1380–1392.e14, doi:10.1016/j.cell.2018.09.037.
- Katada, T. The Inhibitory G Protein G(i) Identified as Pertussis Toxin-Catalyzed ADP-Ribosylation. Biol. Pharm. Bull. 2012, 35, 2103–2111, doi:10.1248/bpb.b212024.
- Littler, D.R.; Ang, S.Y.; Moriel, D.G.; Kocan, M.; Kleifeld, O.; Johnson, M.D.; Tran, M.T.; Paton, A.W.; Paton, J.C.; Summers, R.J.; et al. Structure-Function Analyses of a Pertussis-like Toxin from Pathogenic Escherichia Coli Reveal a Distinct Mecha-nism of Inhibition of Trimeric G-Proteins. J. Biol. Chem. 2017, 292, 15143–15158, doi:10.1074/jbc.M117.796094.
- Qiu, J.; Sheedlo, M.J.; Yu, K.; Tan, Y.; Nakayasu, E.S.; Das, C.; Liu, X.; Luo, Z.-Q. Ubiquitination Independent of E1 and E2 Enzymes by Bacterial Effectors. Nature 2016, 533, 120–124, doi:10.1038/nature17657.
- Bhogaraju, S.; Kalayil, S.; Liu, Y.; Bonn, F.; Colby, T.; Matic, I.; Dikic, I. Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell 2016, 167, 1636–1649.e13, doi:10.1016/j.cell.2016.11.019.
- Yan, F.; Huang, C.; Wang, X.; Tan, J.; Cheng, S.; Wan, M.; Wang, Z.; Wang, S.; Luo, S.; Li, A.; et al. Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination. Mol. Cell 2020, 78, 641–652.e9, doi:10.1016/j.molcel.2020.03.016.
- Watanabe, M.; Kono, T.; Matsushima-Hibiya, Y.; Kanazawa, T.; Nishisaka, N.; Kishimoto, T.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Molecular Cloning of an Apoptosis-Inducing Protein, Pierisin, from Cabbage Butterfly: Possible In-volvement of ADP-Ribosylation in Its Activity. Proc. Natl. Acad. Sci. USA 1999, 96, 10608–10613, doi:10.1073/pnas.96.19.10608.
- Takamura-Enya, T.; Watanabe, M.; Totsuka, Y.; Kanazawa, T.; Matsushima-Hibiya, Y.; Koyama, K.; Sugimura, T.; Waka-bayashi, K. Mono(ADP-Ribosyl)Ation of 2’-Deoxyguanosine Residue in DNA by an Apoptosis-Inducing Protein, Pierisin-1, from Cabbage Butterfly. Proc. Natl. Acad. Sci. USA 2001, 98, 12414–12419, doi:10.1073/pnas.221444598.
- Watanabe, M.; Kono, T.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Purification of Pierisin, an Inducer of Apoptosis in Human Gastric Carcinoma Cells, from Cabbage Butterfly, Pieris Rapae. Jpn. J. Cancer Res. Gann 1998, 89, 556–561, doi:10.1111/j.1349-7006.1998.tb03297.x.
- Nakano, T.; Matsushima-Hibiya, Y.; Yamamoto, M.; Takahashi-Nakaguchi, A.; Fukuda, H.; Ono, M.; Takamura-Enya, T.; Kinashi, H.; Totsuka, Y. ADP-Ribosylation of Guanosine by SCO5461 Protein Secreted from Streptomyces Coelicolor. Toxi-con Off. J. Int. Soc. Toxinology 2013, 63, 55–63, doi:10.1016/j.toxicon.2012.11.019.
- Lyons, B.; Ravulapalli, R.; Lanoue, J.; Lugo, M.R.; Dutta, D.; Carlin, S.; Merrill, A.R. Scabin, a Novel DNA-Acting ADP-Ribosyltransferase from Streptomyces Scabies. J. Biol. Chem. 2016, 291, 11198–11215, doi:10.1074/jbc.M115.707653.
- Schirmer, J.; Wieden, H.-J.; Rodnina, M.V.; Aktories, K. Inactivation of the Elongation Factor Tu by Mosquitocidal Tox-in-Catalyzed Mono-ADP-Ribosylation. Appl. Environ. Microbiol. 2002, 68, 4894–4899, doi:10.1128/aem.68.10.4894-4899.2002.
- Wolf, P.; Elsässer-Beile, U. Pseudomonas Exotoxin A: From Virulence Factor to Anti-Cancer Agent. Int. J. Med. Microbiol. IJMM 2009, 299, 161–176, doi:10.1016/j.ijmm.2008.08.003.
- Oppenheimer, N.J.; Bodley, J.W. Diphtheria Toxin. Site and Configuration of ADP-Ribosylation of Diphthamide in Elonga-tion Factor 2. J. Biol. Chem. 1981, 256, 8579–8581.
- Jørgensen, R.; Purdy, A.E.; Fieldhouse, R.J.; Kimber, M.S.; Bartlett, D.H.; Merrill, A.R. Cholix Toxin, a Novel ADP-Ribosylating Factor from Vibrio Cholerae. J. Biol. Chem. 2008, 283, 10671–10678, doi:10.1074/jbc.M710008200.
- Baysarowich, J.; Koteva, K.; Hughes, D.W.; Ejim, L.; Griffiths, E.; Zhang, K.; Junop, M.; Wright, G.D. Rifamycin Antibiotic Resistance by ADP-Ribosylation: Structure and Diversity of Arr. Proc. Natl. Acad. Sci. USA 2008, 105, 4886–4891, doi:10.1073/pnas.0711939105.
- Gibson, B.A.; Kraus, W.L. New Insights into the Molecular and Cellular Functions of Poly(ADP-Ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424, doi:10.1038/nrm3376.
- Suskiewicz, M.J.; Palazzo, L.; Hughes, R.; Ahel, I. Progress and Outlook in Studying the Substrate Specificities of PARPs and Related Enzymes. FEBS J. 2020, doi:10.1111/febs.15518.
- Rack, J.G.M.; Palazzo, L.; Ahel, I. (ADP-Ribosyl)Hydrolases: Structure, Function, and Biology. Genes Dev. 2020, 34, 263–284, doi:10.1101/gad.334631.119.
