Satellite DNA (satDNA) is defined as highly repetitive DNA consisting of short sequences tandemly repeated a large number of times. Collectively known as the "satellitome", this genomic component offers exciting evolutionary insights into aspects of primate genome biology that raise new questions and challenge existing paradigms.
- satellite DNA
- DNA repeats
- genome
- evolution
- primates
1. Introduction
Primate genomes are enriched in repeats (more than 50%), some of which remain uncharacterized[1][2] [18,19]. Similar to other vertebrates, primate genomes include an abundance of tandem repeats that are organized in such a pattern that the sequences are repeated directly adjacent to each other[3] [20]. These repeat sequences consist of satellite DNA (satDNA), which is defined as tandemly arranged repeats that represent a considerable proportion of the heterochromatic portion of the eukaryotic genome, forming the main structural component (heterochromatin) of chromosomes [4][5][6][7][8][9][10][13,21,22,23,24,25,26]. SatDNA has been implicated in a variety of important functions, including segregation during cell division, homologous chromosomal pairing, kinetochore formation, chromatid attachment, chromosomal rearrangements, and differentiation of sex chromosomes[11][12][13][14][15][16][17] [27,28,29,30,31,32,33]. Perhaps most importantly, satDNA can constitute rapidly evolving sequences of the genome[18][19][20] [34,35,36] and is now considered to be important in driving genomic and karyotypic evolution[4][6][7][8][21] [13,22,23,24,37].
A range of evidence have been collected to propose the dynamics of satDNA in primates which can be characterized as follows: (i) SatDNA repeats may follow an independent evolution in primate genomes and differences in their genomic abundance among taxa can increase with phylogenetic distance, (ii) the predominant satDNA families are conserved in primates with the exception of certain satDNA types that have undergone extreme divergence, (iii) specific portions of satDNA in the genome show population/species/lineage-level divergence and a paradoxical link with the evolution of centromeres, (iv) the Library model of satDNA evolution is still applicable in primate genome, and (v) satDNA transcriptional activity can mediate regulation of gene expressions that consequently influence wide ranging cellular phenomena.
2. Satellite DNA Abundance in Different Primate Lineages
The genomes of most primates, such as monkeys, apes, and humans, comprise up to 50% repeat contents, of which satDNA may constitute as much as 10% of the total number of repeats [22][23][50,51]. RepeatMasker data[24] [52] for different primate species indicate that their genomes can contain a highly variable proportion of satDNA (Figure 1). Comparison of these data shows that satellite repeats are highly abundant in certain families, such as nocturnal primates (superfamily Lorisoidea), strepsirrhine primates (family Cheirogaleidae), and haplorrhine primates (family Tarsiidae) (Figure 1), which suggests extensive expansion of satDNA in the genomes of these lineages. By contrast, in Hominidae and Hylobatidae, satellite repeats are comparatively low in abundance. The genomes of Hominidae and Hylobatidae are invaded by TEs at higher proportions compared with those of the Lorisoidea, Cheirogaleidae, and Tarsiidae lineages. This observation suggests that phylogenetically close lineages show similar patterns of satellite abundance in their genomes, whereas differences in abundance among taxa increase with phylogenetic distance. However, data on the relative percentage of satDNA in different primate genomes must be treated carefully, and precise information on satDNA abundance in primate genomes is still lacking due to misassemblies, gaps, and unresolved assembled centromeric regions that span these repeats [2][25][19,55].
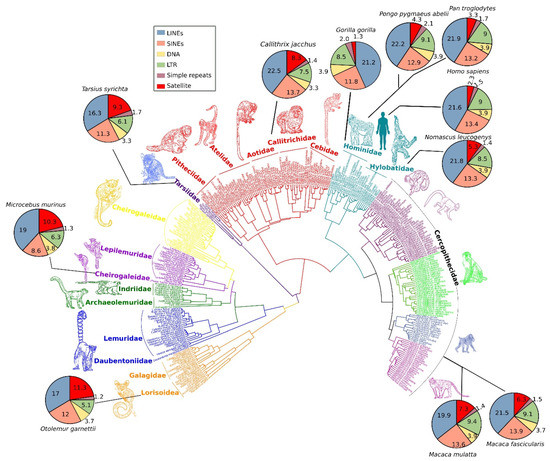
Figure 1. A comprehensive phylogeny of 301 primate species based on mitochondrial DNA sequences using Bayesian inference. Pie charts for selected common primate species show percentage differences of repeat types in the respective genomes. The abundance of satellite DNA in primate genomes varies considerably among lineages (red colored area of pie charts). Additionally, the comparative repeatomic landscape shows LINEs and SINEs emerged as the most expanded elements of primate genomes (blue- and orange-colored areas of pie charts) with consistent pattern across diverse lineages.
SatDNAs were initially identified by their buoyant densities (in g/mL) on cesium chloride gradients[26] [56]. This technique was formerly employed for satDNA detection and biased procedures. This technique can identify a single satellite or sometimes multiple satellites in a genome but cannot detect the entire set of satellite families. Modern techniques, such as NGS and fluorescence in situ hybridization (FISH), have replaced traditional methods and have substantially improved detection and characterization of satDNA[2] [19]. This methodological shift has brought advances in the identification of different repeat types and structural units of satDNA in primate genomes[27][28] [41,47]. Using cytogenetics, the genomic organization and diversity of satDNA have been widely studied mostly in humans, and to some extent in other primate genomes. As a result, a wealth of knowledge is now available on the localizations of satDNA repeats, their lengths, and different units, variability, and number of copies in different genomes[29][30][31][32][33][34][35] [55,57,58,59,60,61,62,63]. These repeats can be categorized as different types of satDNA to better understand their roles, evolution, and applications in phylogenetic analyses. This can include satellites that are generally shared across all eukaryotic lineages and those that are exclusive to primate genomes.
3. General and Primate-Specific satDNA Types
Certain tandem repeat sequences can be classified by the number of base pairs (bp) into two types as microsatellites (ranging in length from one to six or more bp) and minisatellites (usually from 10 to 100 bp)[36] [64]. The human genome contains as much as 3% microsatellites [37][65] and several thousand chromosomal loci enriched with minisatellites[38] [66], also called variable number tandem repeats (VNTRs)[39] [67]. Previous isolation of microsatellites from the human genome has enabled researchers to amplify these sequences in several NHP species, including apes, baboons, macaques, and some platyrrhine monkeys[39][40][41][42][43][44][45][46] [68,69,70,71,72,73,74]. Microsatellites tend to accumulate many substitutions and/or insertions/deletions, and are thus considered to show limited conservation across primate lineages[47] [75]. Many conserved microsatellites, such as AP74, which was discovered in New World monkeys, exhibit similar sequence length (up to 176 bp) in monkeys and humans[48][49] [76,77]. Boán et al.[50] [78] identified the minisatellite MsH42 in the human genome and performed a comparative analysis in 11 NHP species. Phylogenetic analysis detected several variants of MsH42 and the evolutionary birth of minisatellites in the primate genome was hypothesized. According to this hypothesis, the evolutionary birth of MsH42 took place within an intron early in primate lineage evolution and more than 40 million years ago. Then, various mutations including insertions, duplications, and single nucleotide polymorphism of repeat blocks were probably the major forces governing the generation of this minisatellite and its divergence throughout primate evolution[50][78]. Certain (TTAGGG)n sequences, which are specific monomers of microsatellites, can be repeated multiple times, eventually forming the bulk of the telomeric region up to 15 kb on human chromosomes[51][52] [79,80]. These telomeric repeats can serve as binding sites for certain nucleoproteins, such as TRF1, TRF2, and POT1, forming a complex termed “shelterin”[53] [81] that interacts with a ribonucleoprotein[54] [82]. This complex is involved in DNA repair processes and the protection against degradation of chromosomal ends[55] [83].
Well-characterized telomeric satellites of the human genome can also be applied broadly as informative markers to study a variety of hominoid species owing to multiallelic variation and a high degree of heterozygosity[41] [70]. The MsH42 locus shows high similarity with immunoglobulin regions and is involved in recombination events as well as in promoting high rates of unequal crossovers [56][57][78,84,85]. The telomeres harbor short stretches of sequences termed interstitial telomeric sequences (ITSs), which are located far from the chromosomal ends. To trace the evolutionary origin of these sequences in NHP genomes, 22 ITS loci from the human genome were compared with their orthologs in 12 NHPs, representing species such as great apes, gibbons, Old World monkeys, and New World monkeys. Comparison of sequences indicated that, unlike other microsatellites, these ITS sequences were not derived from expansion of pre-existing TTAGGG monomers but rather emerged abruptly during genome evolution in primates as a result of double-strand break repair [58][86]. Similar findings were observed from investigation of a chimpanzee-specific ITS. A universal satDNA classification is still the subject of debate; however, most commonly, satDNA can be grouped according to position and association with different chromosomal loci. SatDNA is primarily clustered within the heterochromatin regions of primate chromosomes. The heterochromatic portion is mainly localized in centromeric and telomeric regions, and sometimes within the interstitial regions of the chromosomes[59] [87], whereas satDNA sequences are mostly located in centromeric regions, and the nearby pericentromeres may be enriched with TEs. Different types of primate satDNA are discussed and summarized as Supplementary Table S1.
3.1. Centromeric and Pericentromeric satDNA: Primate-Specific Alpha Satellites and HORS
The centromere cores of human chromosomes span abundant and highly enriched stretches of satDNA, and are surrounded by heterochromatin containing a combination of short satDNA sequences and retroelements[13] [60][29,88]. Occasionally, these centromeric regions are termed “satellite centromeres”[61] [89]. The centromere is an important region of the chromosome for preservation of genetic materials and plays a critical role in chromosome segregation, cell division, kinetochore organization, and spindle attachment[61][62][63][64] [89,90,91,92]. In primates, the bulk of the centromere is composed of the pancentromeric alpha satellite (AS), organized as stretches of 171 bp monomers in a head-to-tail fashion extending for ~250 kbp up to ~5 Mbp per chromosome [65][66][67][68][93,94,95,96] (Figure 3a(i)). This structure has been reported across diverse groups, including great apes, Old World monkeys, and New World monkeys [69][70][71][72][73][74[96,97,98,99,100,101,102]. These centromere-associated satellites are arranged as superfamilies (SFs) that can be orthologous between human and gorilla[32] [60]. The surrounding pericentromeric satDNA are essential elements that assist in stabilization of DNA–protein binding and regulation of chromosome segregation[75][76] [58,61]. These pericentromeric satellites vary greatly across NHP species but can be conserved among closely related species or may be species-specific[3][7775] [20,103]. For instance, a large block of human chromosome 9 that spans a pericentromeric area enriched with satellite III (SatIII) shares close homology with the gorilla sequence [7876][104]. The Y chromosome of NHPs may carry higher numbers of copies of satellite III sequences than the human Y chromosome [7977][105]. FISH mapping of the pericentromeric-type satellite pW-1 SatIII DNA on chromosomes of various NHP species showed that these sequences might be lacking in the genomes of squirrel monkey (Saimiri sciureus) and baboon (Papio hamadryas)[7977] [105]. These centromeric satellites can vary substantially across different species, but certain species-specific or even highly conserved satDNA may also be present in the centromere domains[3][7775][20,103]. For example, two major families of centromeric satellites, termed C1 and C2, detected in Old World monkey species crested mona monkey (Cercopithecus pogonias) and sun-tailed monkey (Cercopithecus solatus) have remained highly conserved[8078] [48]. For Old World monkeys, apes, and humans, each genome harbors evolutionarily distinct AS monomers[8179] [106]. Although most primate centromeres can be enriched with satellites repeats, there are certain chromosomes of orangutan that comprise non-repeated centromeres[64][8280][8381][8482] [92,107,108,109,110]. In such cases, the centromeres may resemble newly formed neocentromeres as a result of disruption in the centromeric region, such as in humans[64][8583][92,111]. Such non-repeated centromeres are likely to be evolutionary new centromeres (ENCs), forming neocentromeres that might have subsequently gained repeat sequences to stabilize the genome and become fixed in populations. This phenomenon can also occur in the centromeres of several non-primate species, such as horse and chicken[8684][8785] [112,113]. In the following, we focus mainly on the predominant centromeric satDNA in primate genomes as AS repeats.
The AS repeats were first observed as tandem repeats in the African green monkey (Chlorocebus aethiops) genome[65] [93], followed by identification of homologous repeats in New World monkeys and apes[68][8886] [96,114]. These sequences are considered to be critical components for the various functions of primate centromeres[66] [94]. Previous results suggest that AS sequences were involved in stabilization of ENCs after their emergence in primate genomes[8381][8987] [109,115]. Human and macaque chromosomes contain a total of 14 ENCs, of which nine ENCs in the macaque genome show abundant arrays of AS[8381] [109]. Interestingly, ENCs occur in macaque chromosome 4 and human chromosome 6, which are orthologous to each other (Figure 2a(ii)) [8381][9088][9189][109,116,117].
The AS monomer size is 171 bp, tandemly arranged in a head-to-tail manner, and shows as much as 70% sequence similarity. The combined monomers can form a long array spanning an uninterrupted 250–5000 kb stretch of repeated satellites, giving rise to high-order repeats (HORs) (Figure 2a(iii)). A certain monomer in the HORs with a sequence size of 17 bp is termed the CENP-B box. This motif acts as a protein-binding site for a centromeric CENP-B protein in primates. The human genome project, which was declared complete in 2003, was still unable to recover a large proportion of the centromeric and other repeats, including more than 10% of the contents of the whole genome, mainly sex chromosomes. However, subsequent technological developments enabled assembly of the entire human Y chromosomal centromere[64][9290] [62,118]. The Y chromosome assembly could be used as a reference sequence to extend evolutionary insights into the centromeric repeats of NHPs for which Y chromosome assemblies have not been hitherto accomplished.
In primates, the flanked regions of centromeres have specialized HORs arrays, whereas AS sequences are organized as non-structured and heterogeneous repeats, forming distinctive pericentromeres. In these pericentromeres, AS sequence repeats are arranged as monomers instead of HORs and are interrupted with additional elements, mainly retrotranposable elements in humans[9391] [119] (Figure 2a(iii)), which may also be common to other primate genomes. The pericentromeres of certain human chromosomes may also show enrichment of several other repeat sequences, including the 5 bp satDNA II and III type sequences[7775][9492] [103,120]. The AS sequences can show nucleotide variation when one monomer is compared with the repeats of the same array, with nucleotide identity ranging from 70% to 90%. The sequences of a monomer in one array may show up to 95% similarity with its counterpart unit in the other array at the same locus [35][9593][9694][63,121,122]. In the human genome, the organization of HORs with their monomer units has been extensively studied[37][69][9795][9896] [65,97,123,124], and shows the occurrence of various subfamilies of chromosome-specific AS sequences. The sequences of HORs in great apes, such as orangutan, gorilla, and chimpanzee, show a lower degree of variation in comparison with HORs observed in the human genome[97][98][99][100][101][102] [125,126,127,128]. Initially, it was presumed that the organization of HORs might be restricted to hominids; however, HORs were subsequently detected in the genomes of gibbons[73][74][103101] [101,102,129] and of Old World and New World monkeys[8078][74[104102] [48,102,130]. During the evolution of the primate genome, the 170 bp AS monomer underwent a series of sequence variations [59][87]. A novel AS monomer type of 189 bp was discovered in the centromeres of gorilla [60]. Chromosome-specific subfamilies are absent in Old World and New World monkeys as well as in gibbons[59][73][8179] [87,101,106]. Cloning, sequencing, and hybridization of acrocentric chromosomes revealed novel AS sequence repeats in Azara’s owl monkey (Aotus azarae), which is a species of New World monkey[7][8] [22,23]. These repeats include three megasatellites, namely OwlRep, OwlAlp1, and OwlAlp2, which vary in size from 184 to 344 bp as identified in the centromeric and pericentromeric regions. Analysis of retina samples using three-dimensional FISH revealed that OwlRep is the major component of heterochromatin, which indicates its role in the evolution of night vision in this species[105103][106104] [131,132]. Recently, Cacheux et al.[107105] [49] investigated the evolutionary dynamics of AS sequence repeats and their diversity in the Old World monkeys Cercopithecus pogonias and C. solatus using targeted sequencing and FISH mapping. These authors reported evidence of chromosome-specific subfamilies that might have evolved through homogenization. The OwlRep repeat shows ~82% homology with a satellite sequence termed HSAT6, which is a 126 bp long tandem centromeric repeat. The HSAT6 sequence was also detected in the owl monkey genome, and comparative analysis revealed its broad distribution among hominoids and New World and Old World monkeys. Phylogenetic analysis confirmed that OwlRep evolved from HSAT6[106104] [132].
In addition to AS, an additional type of satellite family termed the beta satellite is distributed in the heterochromatin of primates[108106][109107][110108] [133,134,135]. Beta satDNA are repeats that comprise ~68 bp monomers. They are predominantly organized in the shorter arm of acrocentric chromosomes and arranged in stretches several kb in length[109][110][111][112][113][114] [136,137,138,139]. The beta satDNA repeats can form complexes with arrays of specific repeats, termed D4Z4 repeats, at certain acrocentric loci, such as 10q26 and 4q35[115113][116114] [140,141]. Evolutionary analyses involving cloning and FISH experiments have predicted that 4q35 containing D4Z4 repeats might represent an ancestral locus with an extensively radiated sequence region that evolved after the divergence of hominoids and Old World monkeys[117115][118116][119117] [142,143,144]. The origin and evolution of beta satDNA vary in diverse species of hominids, such as humans, chimpanzee, and gorilla[120118][121119] [145,146]. FISH mapping data confirm that D4Z4 is also conserved in Old World and New World monkeys, whereas in primates distantly related to humans (e.g., lemurs), this sequence has retained tandem repetition but conservation is limited to promotor regions[122120] [147]. Genomic analysis of orangutan has revealed the origin of beta satDNA in earlier ancestors of hominoids and shows that these repeats are preferentially located in pericentromeres[110108] [135]. This study concluded that these repeats originated as low copies, remained non-duplicated in the early ape ancestors, and later evolved as duplicons acquiring the typical characteristics of classical satellites in humans and other primates. Adjacent to ASs, the classical non-alphoid satDNA repeat families I, II, and III are located in pericentromeres of human chromosomes[67] [95]. The human genome includes the Sat III family, which is composed of GGAAT and GGAGT repeat sequences in different percentages. The satellite III family is mainly localized on the short arm of acrocentric chromosomes in humans and other primate species. This family is also present in the chimpanzee, gorilla, and orangutan genomes[123121] [124122][148,149]. The chromosomal organization of this satellite family has provided interesting evolutionary insights into primate genomes[79] [105]. Sequence comparisons have detected variation across different primate species and suggest that the Sat III family might have appeared ~16–23 million years ago in Hominoidea[79] [105]. The evolutionary origin and extensive diversification of centromeric satellites in primate genomes remain unclear; however, it is speculated that TEs are the possible progenitors and sources that form novel satellites by insertions into existing satellite regions[93] [119].
3.2. Telomeric and Subtelomeric satDNA
The telomere is located at the end of the chromosome and is enriched with a non-coding, repetitive DNA sequence. The 500 kb region of each chromosomal arm terminal is the so-called subtelomeric region[125] [150]. Both telomere and subtelomere have high-density of satDNA repeats. Telomeric regions of the primate genome show a high frequency of minisatellites, which also occur in other loci of chromosomes[126][127] [67,151]. The bulk of telomeric-specific regions are mainly composed of (TTAGGG)n microsatellites in humans[128] [79]. Adjacent to the telomere, the subtelomere region is mostly enriched in rapidly evolving satellite repeats with variable levels of repetitiveness and size [30][129][130][57,152,153]. Although these subtelomeric satellites can be species-specific and often chromosome-specific, there are also satellites that remain highly conserved[131] [154]. The microsatellites (CCCTAA)n, (CCCCAA)n, and (CCCTCA)n are present in telomeres of primates [132][155], whereas (CCCGAA)n is restricted to subtelomeres [133][156] (Figure 3b). In New World monkeys, the subtelomeres can carry novel satDNA sequences. The subtelomeric regions of callitrichid monkeys harbor a satellite termed MarmoSAT that is composed of a 171 bp motif [134][157]. The MarmoSAT occurs as a monomer, whereas in common marmoset (Callithrix jacchus) it is organized in HORs with a sequence of 338 bp. Recently, some intriguing groups of satDNA sequences enriched with AT nucleotides, termed StSats, have been reported in telomeres of humans and great apes, including bonobo, chimpanzee, gorilla, and orangutan[135] [47]. The StSats are located in proximity to telomeric regions [136][137][138][158,159,160]. Astonishingly, these satellites are very highly enriched in the gorilla and chimpanzee genomes compared with their abundance in humans[135] [47]. Previously, it was hypothesized that these repeats occurred in hominid ancestors and were lost in humans[136][137][138] [158,159,160]. The abundance of StSats repeats in the bonobo, chimpanzee, and gorilla genomes indicates that these sequences might contribute to important genomic functions in these species. Different functions have been proposed for these repeats that include their role in meiosis, telomere clustering, and control of replication duration with telomeric regions[136][137][138] [158,159,160].
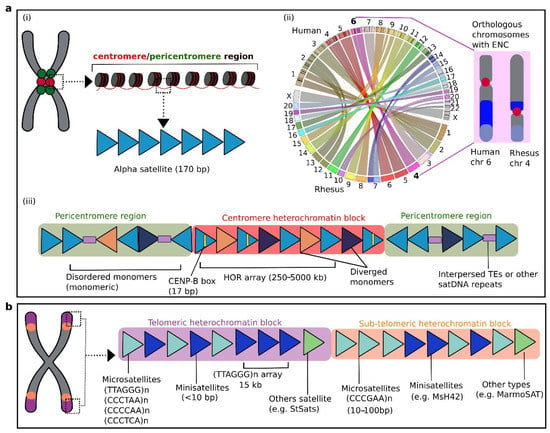
Figure 2. Schematic illustration of satellite DNA repeats and their organization in primate genomes. (a) (i) Primate centromeric (red) and pericentromeric (green) regions are enriched with alpha satellite (AS) DNA as the most abundant satellite repeats of primate genomes and form the bulk of the heterochromatin core. (ii) A sketch highlighting the orthologous chromosomes and centromeric repositioning as evolutionary new centromeres (ENCs) between human and rhesus macaque. The circos plot depicts the syntenic relationship between the two genomes. Circos graphics was plotted using Synteny Portal [117]. Note that human chromosome 6 is completely orthologous to macaque chromosome 4, with evolved centromeres [139][140][109,116]. (iii) The AS constitute the tandem repeat units (blue triangles) and can be either organized as disordered arrays (monomeric) mostly located in pericentromeres, or highly ordered in a head-to-tail fashion (HORs) forming longer arrays in centromeres. Some monomers may also have a short sequence termed the CENP-B box (yellow line), which binds the centromeric regions to the DNA-binding proteins. Diverged monomers (orange and dark triangles), and interspersed repeats (purple rectangles) are also depicted. (b) Telomeric and subtelomeric regions of primate chromosomes are enriched with distinct microsatellites (light blue) and minisatellites (dark blue). Various primate-associated satellite examples are shown.
The distribution of two distinct satellite repeats, termed Cap-A and Cap-B, was reported in a New World monkey species, Cebus paella[141] [161]. The Cap-A sequence is 1500 bp long and forms heterochromatic blocks in the interstitial sites of chromosome 11 and a few telomeric regions. This suggests that this sequence underwent a new episode of amplification in New World monkeys. This satDNA repeat is absent in most marmoset species and present in species of the family Cebidae. By contrast, the Cap-B satellite, which is 342 bp long, is mainly localized in the centromeric regions of many chromosomes of New World monkeys. The Cap-B monomer sequence shares more than 60% identity with AS repeats, which indicates that Cap-B might be the New World monkey homolog of Old World monkey AS repeat sequences. Telomeric satDNA sequences can participate in the formation and maintenance of telomeres, and may have an incidental role in cases losing of conventional telomeric repeats. In this way, telomeric ends are stabilized by satDNA[142] [162]. Further, it has been demonstrated that telomere-like sequences interspersed within subtelomeric DNA may also play a role in subtelomeric recombination and transcription, via alternative lengthening of the telomere pathway and in telomere healing[143] [163]. It is necessary to identify and characterize the telomeric/centromeric satDNA sequences particularly at the breakpoint sites because of their role in mediating chromosomal rearrangements[144] [164] that occurred during primate evolution. Such analyses have been performed with regard to the gorilla-specific translocation[145] [165] as well as the chromosome scale variations that serve to distinguish human and chimpanzee chromosomes. Various hotspot rearrangement regions of the gibbon genome have also been characterized[146] [166]. In contrast to the great apes, gibbons have chromosomes with higher levels of rearrangement compared to ancestral primate karyotypes[147] [167]. A comparison of human and chimpanzee karyotypes showed that two ancestral chromosomal homologs of chimpanzee chromosomes 12 and 13 underwent a fusion event to give rise to human chromosome 2[148] [168]. This fusion was mediated by recombination between telomeric satDNA repeats of the two sub-metacentric ancestral chromosomes. The hyper-expanded repeats are localized in subtelomeric regions of chimpanzee chromosomes. These repeat enriched regions are also prone to other types of rearrangement events such as duplicative transpositions and inter-chromosomal sequence variations[149] [169]. Since many primate-specific rearranged loci are enriched with high-copy repetitive sequence elements such as alpha satDNA repeats, SINEs, LINEs, and LTRs, a range of different molecular mechanisms were probably involved in promoting chromosomal breakage during the evolution of primate genomes[144] [164]. Genome-wide scale analyses at higher resolution are necessary to determine the precise mechanisms underlying the different types of rearrangement, and to assess their relative contribution to the process of evolutionary change.
References
- Marques-Bonet, T.; Ryder, O.A.; Eichler, E.E. Sequencing primate genomes: What have we learned? Annu. Rev. Genomics Hum. Genet. 2009, 10, 355–386.
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2012, 13, 36–46.
- Melters, D.P.; Bradnam, K.R.; Young, H.A.; Telis, N.; May, M.R.; Ruby, J.G.; Sebra, R.; Peluso, P.; Eid, J.; Rank, D.; et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 2013, 14, R10.
- Ahmad, S.F.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of paradigm shift with repeat landscapes in reptiles: Powerful facilitators of chromosomal rearrangements for diversity and evolution (running title: Genomic impact of repeats on chromosomal dynamics in reptiles). Genes 2020, 11, 827.
- Charlesworth, B.; Jarne, P.; Assimacopoulos, S. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. III. Element abundances in heterochromatin. Genet. Res. 1994, 64, 183–197.
- Prakhongcheep, O.; Hirai, Y.; Hara, T.; Srikulnath, K.; Hirai, H.; Koga, A. Two types of alpha satellite DNA in distinct chromosomal locations in Azara’s owl monkey. DNA Res. 2013, 20, 235–240.
- Prakhongcheep, O.; Chaiprasertsri, N.; Terada, S.; Hirai, Y.; Srikulnath, K.; Hirai, H.; Koga, A. Heterochromatin blocks constituting the entire short arms of acrocentric chromosomes of Azara’s owl monkey: Formation processes inferred from chromosomal locations. DNA Res. 2013, 20, 461–470.
- Prakhongcheep, O.; Thapana, W.; Suntronpong, A.; Singchat, W.; Pattanatanang, K.; Phatcharakullawarawat, R.; Muangmai, N.; Peyachoknagul, S.; Matsubara, K.; Ezaz, T.; et al. Lack of satellite DNA species-specific homogenization and relationship to chromosomal rearrangements in monitor lizards (Varanidae, Squamata). BMC Evol. Biol. 2017, 17, 193.
- Thongchum, R.; Singchat, W.; Laopichienpong, N.; Tawichasri, P.; Kraichak, E.; Prakhongcheep, O.; Sillapaprayoon, S.; Muangmai, N.; Baicharoen, S.; Suntrarachun, S.; et al. Diversity of PBI-DdeI satellite DNA in snakes correlates with rapid independent evolution and different functional roles. Sci. Rep. 2019, 9, 15459.
- Suntronpong, A.; Singchat, W.; Kruasuwan, W.; Prakhongcheep, O.; Sillapaprayoon, S.; Muangmai, N.; Somyong, S.; Indananda, C.; Kraichak, E.; Peyachoknagul, S.; et al. Characterization of centromeric satellite DNAs (MALREP) in the Asian swamp eel (Monopterus albus) suggests the possible origin of repeats from transposable elements. Genomics 2020, 112, 3097–3107.
- Nakagawa, T.; Okita, A.K. Transcriptional silencing of centromere repeats by heterochromatin safeguards chromosome integrity. Curr. Genet. 2019, 65, 1089–1098.
- Kim, J.H.; Ebersole, T.; Kouprina, N.; Noskov, V.N.; Ohzeki, J.I.; Masumoto, H.; Mravinac, B.; Sullivan, B.A.; Pavlicek, A.; Dovat, S.; et al. Human gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing. Genome Res. 2009, 19, 533–544.
- Schueler, M.G.; Higgins, A.W.; Rudd, M.K.; Gustashaw, K.; Willard, H.F. Genomic and genetic definition of a functional human centromere. Science 2001, 294, 109–115.
- Schueler, M.G.; Sullivan, B.A. Structural and functional dynamics of human centromeric chromatin. Annu. Rev. Genom. Hum. Genet. 2006, 7, 301–313.
- Aldrup-MacDonald, M.E.; Sullivan, B.A. The past, present, and future of human centromere genomics. Genes 2014, 5, 33–50.
- Fachinetti, D.; Han, J.S.; McMahon, M.A.; Ly, P.; Abdullah, A.; Wong, A.J.; Cleveland, D.W. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev. Cell 2015, 33, 314–327.
- McNulty, S.M.; Sullivan, B.A. Alpha satellite DNA biology: Finding function in the recesses of the genome. Chromosom. Res. 2018, 26, 115–138.
- Jagannathan, M.; Warsinger-Pepe, N.; Watase, G.J.; Yamashita, Y.M. Comparative analysis of satellite DNA in the drosophila melanogaster species complex. G3 Genes Genomes Genet. 2017, 7, 693–704.
- Lower, S.S.; McGurk, M.P.; Clark, A.G.; Barbash, D.A. Satellite DNA evolution: Old ideas, new approaches. Curr. Opin. Genet. Dev. 2018, 49, 70–78.
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230.
- Hartley, G.; O’neill, R.J. Centromere repeats: Hidden gems of the genome. Genes 2019, 10, 223.
- Sullivan, L.L.; Chew, K.; Sullivan, B.A. α satellite DNA variation and function of the human centromere. Nucleus 2017, 8, 331–339.
- Sullivan, L.L.; Sullivan, B.A. Genomic and functional variation of human centromeres. Exp. Cell Res. 2020, 389, 111896.
- Smit, A.; Hubley, R.; Grenn, P. RepeatMasker Open-4.0. 2015. Available online: http://www.repeatmasker.org/ (accessed on 1 August 2020).
- Miga, K.H. Completing the human genome: The progress and challenge of satellite DNA assembly. Chromosom. Res. 2015, 23, 421–426.
- López-Flores, I.; Garrido-Ramos, M.A. The repetitive DNA content of eukaryotic genomes. Genome Dyn. 2012, 7, 1–28.
- Cordaux, R.; Sen, S.K.; Konkel, M.K.; Batzer, M.A. Computational methods for the analysis of primate mobile elements. Methods Mol. Biol. 2010, 628, 137–151.
- Cechova, M.; Harris, R.S.; Tomaszkiewicz, M.; Arbeithuber, B.; Chiaromonte, F.; Makova, K.D. High Satellite Repeat Turnover in Great Apes Studied with Short- And Long-Read Technologies. Mol. Biol. Evol. 2019, 36, 2415–2431.
- Waring, M.; Britten, R.J. Nucleotide sequence repetition: A rapidly reassociating fraction of mouse DNA. Science 1966, 154, 791–794.
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of tandemly repetitive DNA: Functional roles. Chromosom. Res. 2015, 23, 463–477.
- Rogers, J.; Mahaney, M.C.; Witte, S.M.; Nair, S.; Newman, D.; Wedel, S.; Rodriguez, L.A.; Rice, K.S.; Slifer, S.H.; Perelygin, A.; et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics 2000, 67, 237–247.
- Catacchio, C.R.; Ragone, R.; Chiatante, G.; Ventura, M. Organization and evolution of Gorilla centromeric DNA from old strategies to new approaches. Sci. Rep. 2015, 5, 14189.
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153.
- Miga, K.H.; Newton, Y.; Jain, M.; Altemose, N.; Willard, H.F.; Kent, E.J. Centromere reference models for human chromosomes X and y satellite arrays. Genome Res. 2014, 24, 697–707.
- Sujiwattanarat, P.; Thapana, W.; Srikulnath, K.; Hirai, Y.; Hirai, H.; Koga, A. Higher-order repeat structure in alpha satellite DNA occurs in New World monkeys and is not confined to hominoids. Sci. Rep. 2015, 5, 10315.
- Richard, G.F.; Pâques, F. Mini- and microsatellite expansions: The recombination connection. EMBO Rep. 2000, 1, 122–126.
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13.
- Ramel, C. Mini- and microsatellites. EHP 1997, 105, 781–789.
- Näslund, K.; Saetre, P.; Von Salomé, J.; Bergström, T.F.; Jareborg, N.; Jazin, E. Genome-wide prediction of human VNTRs. Genomics 2005, 85, 24–35.
- Blanquer-Maumont, A.; Crouau-Roy, B. Polymorphism, monomorphism, and sequences in conserved microsatellites in primate species. J. Mol. Evol. 1995, 41, 492–497.
- Garza, J.C.; Slatkin, M.; Freimer, N.B. Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size. Mol. Biol. Evol. 1995, 12, 594–603.
- Coote, T.; Bruford, M.W. Human Microsatellites Applicable for Analysis of Genetic Variation in Apes and Old World Monkeys. J. Hered. 1996, 87, 406–410.
- Kayser, M.; Caglià, A.; Corach, D.; Fretwell, N.; Gehrig, C.; Graziosi, G.; Heidorn, F.; Herrmann, S.; Herzog, B.; Hidding, M.; et al. Evaluation of Y-chromosomal STRs: A multicenter study. Int. J. Legal Med. 1997, 110, 125–133.
- Goossens, B.; Chikhi, L.; Utami, S.S.; De Ruiter, J.; Bruford, M.W. A multi-samples, multi-extracts approach for microsatellite analysis of faecal samples in an arboreal ape. Conserv. Genet. 2000, 1, 157–162.
- Nair, S.; Ha, J.; Rogers, J. Nineteen new microsatellite DNA polymorphisms in pigtailed macaques (Macaca nemestrina). Primates 2000, 41, 343–350.
- Winkler, L.A.; Zhang, X.; Ferrell, R.; Wagner, R.; Dahl, J.; Peter, G.; Sohn, R. Geographic Microsatellite Variability in Central American Howling Monkeys. Int. J. Primatol. 2004, 25, 197–210.
- Clisson, I.; Lathuilliere, M.; Crouau-Roy, B. Conservation and evolution of microsatellite loci in primate taxa. Am. J. Primatol. 2000, 50, 205–214.
- Buschiazzo, E.; Gemmell, N.J. Conservation of human microsatellites across 450 million years of evolution. Genome Biol. Evol. 2010, 2, 153–165.
- Oklander, L.I.; Steinberg, E.R.; Mudry, M.D. A new world monkey microsatellite (AP74) higly conserved in primates. Acta Biol. Colomb. 2012, 17, 93–101.
- Boán, F.; Blanco, M.G.; Quinteiro, J.; Mouriño, S.; Gómez-Márquez, J. Birth and Evolutionary History of a Human Minisatellite. Mol. Biol. Evol. 2004, 21, 228–235.
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)(n), present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626.
- O’Sullivan, R.J.; Karlseder, J. Telomeres: Protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010, 11, 171–181.
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin protects chromosome ends by compacting telomeric chromatin. Cell 2016, 164, 735–746.
- Wyatt, H.D.M.; West, S.C.; Beattie, T.L. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010, 38, 5609–5622.
- Maddar, H.; Ratzkovsky, N.; Krauskopf, A. Role for telomere cap structure in meiosis. Mol. Biol. Cell 2001, 12, 3191–3203.
- Boán, F.; Rodríguez, J.M.; Gómez-Márquez, J. A non-hypervariable human minisatellite strongly stimulates in vitro intramolecular homologous recombination. J. Mol. Biol. 1998, 278, 499–505.
- Boán, F.; Rodríguez, J.M.; Mouriño, S.; Blanco, M.G.; Viñas, A.; Sánchez, L.; Gómez-Márquez, J. Recombination analysis of the human minisatellite MsH42 suggests the existence of two distinct pathways for initiation and resolution of recombination at MsH42 in rat testes nuclear extracts. Biochemistry 2002, 41, 2166–2176.
- Nergadze, S.G.; Rocchi, M.; Azzalin, C.M.; Mondello, C.; Giulotto, E. Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Res. 2004, 14, 1704–1710.
- Plohl, M.; Meštrović, N.; Mravinac, B. Satellite DNA evolution. Genome Dyn. 2012, 7, 126–152.
- Kazakov, A.E.; Shepelev, V.A.; Tumeneva, I.G.; Alexandrov, A.A.; Yurov, Y.B.; Alexandrov, I.A. Interspersed repeats are found predominantly in the “old” α satellite families. Genomics 2003, 82, 619–627.
- Steiner, F.A.; Henikoff, S. Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev. 2015, 31, 28–35.
- Plohl, M.; Luchetti, A.; Meštrović, N.; Mantovani, B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 2008, 409, 72–82.
- Verdaasdonk, J.S.; Bloom, K. Centromeres: Unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 2011, 12, 320–332.
- Fukagawa, T.; Earnshaw, W.C. The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 2014, 30, 496–508.
- Maio, J.J. DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J. Mol. Biol. 1971, 56, 579–595.
- Manuelidis, L.; Wu, J.C. Homology between human and simian repeated DNA. Nature 1978, 276, 92–94.
- Vissel, B.; Andy Choo, K.H. Evolutionary relationships of multiple alpha satellite subfamilies in the centromeres of human chromosomes 13, 14, and 21. J. Mol. Evol. 1992, 35, 137–146.
- Musich, P.R.; Brown, F.L.; Maio, J.J. Highly repetitive component α and related alphoid DNAs in man and monkeys. Chromosoma 1980, 80, 331–348.
- Willard, H.F.; Waye, J.S. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 1987, 3, 192–198.
- Alves, G.; Seuánez, H.N.; Fanning, T. Alpha satellite DNA in neotropical primates (Platyrrhini). Chromosoma 1994, 103, 262–267.
- Alves, G.; Canavez, F.; Seuánez, H.; Fanning, T. Recently amplified satellite DNA in Callithrix argentata (Primates, Platyrrhini). Chromosom. Res. 1995, 3, 207–213.
- Alexandrov, I.; Kazakov, A.; Tumeneva, I.; Shepelev, V.; Yurov, Y. Alpha-satellite DNA of primates: Old and new families. Chromosoma 2001, 110, 253–266.
- Cellamare, A.; Catacchio, C.R.; Alkan, C.; Giannuzzi, G.; Antonacci, F.; Cardone, M.F.; Della Valle, G.; Malig, M.; Rocchi, M.; Eichler, E.E.; et al. New insights into centromere organization and evolution from the white-cheeked Gibbon and marmoset. Mol. Biol. Evol. 2009, 26, 1889–1900.
- Akihiko, K.; Yuriko, H.; Shoko, T.; Israt, J.; Sudarath, B.; Visit, A.; Hirohisa, H. Evolutionary origin of higher-order repeat structure in alpha-satellite DNA of primate centromeres. DNA Res. 2014, 21, 407–415.
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of tandemly repetitive DNA: Functional roles. Chromosom. Res. 2015, 23, 463–477, doi:10.1007/s10577-015-9494-4.Plohl, M.; Meštrović, N.; Mravinac, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325.
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153, doi:10.1073/pnas.1518008112.Pita, M.; Gosálvez, J.; Gosálvez, A.; Nieddu, M.; López-Fernández, C.; Mezzanotte, R. A highly conserved pericentromeric domain in human and gorilla chromosomes. Cytogenet. Genome Res. 2010, 126, 253–258.
- Plohl, M.; Meštrović, N.; Mravinac, B. Centromere identity from the DNA point of view. Chromosoma 2014, 123, 313–325.Jarmuz, M.; Glotzbach, C.D.; Bailey, K.A.; Bandyopadhyay, R.; Shaffer, L.G. The evolution of satellite III DNA subfamilies among primates. Am. J. Hum. Genet. 2007, 80, 495–501.
- Pita, M.; Gosálvez, J.; Gosálvez, A.; Nieddu, M.; López-Fernández, C.; Mezzanotte, R. A highly conserved pericentromeric domain in human and gorilla chromosomes. Cytogenet. Genome Res. 2010, 126, 253–258.Cacheux, L.; Ponger, L.; Gerbault-Seureau, M.; Richard, F.A.; Escudé, C. Diversity and distribution of alpha satellite DNA in the genome of an Old World monkey: Cercopithecus solatus. BMC Genom. 2016, 17, 916.
- Jarmuz, M.; Glotzbach, C.D.; Bailey, K.A.; Bandyopadhyay, R.; Shaffer, L.G. The evolution of satellite III DNA subfamilies among primates. Am. J. Hum. Genet. 2007, 80, 495–501.Alkan, C.; Ventura, M.; Archidiacono, N.; Rocchi, M.; Sahinalp, S.C.; Eichler, E.E. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput. Biol. 2007, 3, 1807–1818.
- Cacheux, L.; Ponger, L.; Gerbault-Seureau, M.; Richard, F.A.; Escudé, C. Diversity and distribution of alpha satellite DNA in the genome of an Old World monkey: Cercopithecus solatus. BMC Genom. 2016, 17, 916.Montefalcone, G.; Tempesta, S.; Rocchi, M.; Archidiacono, N. Centromere repositioning. Genome Res. 1999, 9, 1184–1188.
- Alkan, C.; Ventura, M.; Archidiacono, N.; Rocchi, M.; Sahinalp, S.C.; Eichler, E.E. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput. Biol. 2007, 3, 1807–1818.Ventura, M.; Antonacci, F.; Cardone, M.F.; Stanyon, R.; D’Addabbo, P.; Cellamare, A.; Sprague, L.J.; Eichler, E.E.; Archidiacono, N.; Rocchi, M. Evolutionary formation of new centromeres in macaque. Science 2007, 316, 243–246.
- Montefalcone, G.; Tempesta, S.; Rocchi, M.; Archidiacono, N. Centromere repositioning. Genome Res. 1999, 9, 1184–1188.Stanyon, R.; Rocchi, M.; Capozzi, O.; Roberto, R.; Misceo, D.; Ventura, M.; Cardone, M.F.; Bigoni, F.; Archidiacono, N. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosom. Res. 2008, 16, 17–39.
- Ventura, M.; Antonacci, F.; Cardone, M.F.; Stanyon, R.; D’Addabbo, P.; Cellamare, A.; Sprague, L.J.; Eichler, E.E.; Archidiacono, N.; Rocchi, M. Evolutionary formation of new centromeres in macaque. Science 2007, 316, 243–246.Amor, D.J.; Andy Choo, K.H. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 2002, 71, 695–714.
- Stanyon, R.; Rocchi, M.; Capozzi, O.; Roberto, R.; Misceo, D.; Ventura, M.; Cardone, M.F.; Bigoni, F.; Archidiacono, N. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chromosom. Res. 2008, 16, 17–39.Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867.
- Amor, D.J.; Andy Choo, K.H. Neocentromeres: Role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 2002, 71, 695–714.Shang, W.H.; Hori, T.; Toyoda, A.; Kato, J.; Popendorf, K.; Sakakibara, Y.; Fujiyama, A.; Fukagawa, T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010, 20, 1219–1228.
- Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867.Maio, J.J.; Brown, F.L.; Musich, P.R. Toward a molecular paleontology of primate genomes—I. The HindIII and EcoRI dimer families of alphoid DNAs. Chromosoma 1981, 83, 103–125.
- Shang, W.H.; Hori, T.; Toyoda, A.; Kato, J.; Popendorf, K.; Sakakibara, Y.; Fujiyama, A.; Fukagawa, T. Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 2010, 20, 1219–1228.Kalitsis, P.; Choo, K.H.A. The evolutionary life cycle of the resilient centromere. Chromosoma 2012, 121, 327–340.
- Maio, J.J.; Brown, F.L.; Musich, P.R. Toward a molecular paleontology of primate genomes—I. The HindIII and EcoRI dimer families of alphoid DNAs. Chromosoma 1981, 83, 103–125.McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29.
- Kalitsis, P.; Choo, K.H.A. The evolutionary life cycle of the resilient centromere. Chromosoma 2012, 121, 327–340.Lee, J.; Hong, W.Y.; Cho, M.; Sim, M.; Lee, D.; Ko, Y.; Kim, J. Synteny Portal: A web-based application portal for synteny block analysis. Nucleic Acids Res. 2016, 44, W35–W40.
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29.Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017, 27, 849–864.
- Lee, J.; Hong, W.Y.; Cho, M.; Sim, M.; Lee, D.; Ko, Y.; Kim, J. Synteny Portal: A web-based application portal for synteny block analysis. Nucleic Acids Res. 2016, 44, W35–W40.Klein, S.J.; O’Neill, R.J. Transposable elements: Genome innovation, chromosome diversity, and centromere conflict. Chromosom. Res. 2018, 26, 5–23.
- Schneider, V.A.; Graves-Lindsay, T.; Howe, K.; Bouk, N.; Chen, H.C.; Kitts, P.A.; Murphy, T.D.; Pruitt, K.D.; Thibaud-Nissen, F.; Albracht, D.; et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017, 27, 849–864.Prosser, J.; Frommer, M.; Paul, C.; Vincent, P.C. Sequence relationships of three human satellite DNAs. J. Mol. Biol. 1986, 187, 145–155.
- Klein, S.J.; O’Neill, R.J. Transposable elements: Genome innovation, chromosome diversity, and centromere conflict. Chromosom. Res. 2018, 26, 5–23.Willard, H.F. Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 1985, 37, 524–532.
- Prosser, J.; Frommer, M.; Paul, C.; Vincent, P.C. Sequence relationships of three human satellite DNAs. J. Mol. Biol. 1986, 187, 145–155.Warburton, P.E.; Willard, H.F. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of α-satellite DNA. J. Mol. Biol. 1990, 216, 3–16.
- Willard, H.F. Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 1985, 37, 524–532.Paar, V.; Basar, I.; Rosandic, M.; Gluncic, M. Consensus Higher Order Repeats and Frequency of String Distributions in Human Genome. Curr. Genom. 2007, 8, 93–111.
- Warburton, P.E.; Willard, H.F. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of α-satellite DNA. J. Mol. Biol. 1990, 216, 3–16.Aldrup-MacDonald, M.E.; Kuo, M.E.; Sullivan, L.L.; Chew, K.; Sullivan, B.A. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genom. Res. 2016, 26, 1301–1311.
- Paar, V.; Basar, I.; Rosandic, M.; Gluncic, M. Consensus Higher Order Repeats and Frequency of String Distributions in Human Genome. Curr. Genom. 2007, 8, 93–111.Willard, H.F. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1991, 1, 509–514.
- Aldrup-MacDonald, M.E.; Kuo, M.E.; Sullivan, L.L.; Chew, K.; Sullivan, B.A. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. Genom. Res. 2016, 26, 1301–1311.Haaf, T.; Warburton, P.E.; Willard, H.F. Integration of human α-satellite DNA into simian chromosomes: Centromere protein binding and disruption of normal chromosome segregation. Cell 1992, 70, 681–696.
- Willard, H.F. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1991, 1, 509–514.Warburton, P.E.; Haaf, T.; Gosden, J.; Lawson, D.; Willard, H.F. Characterization of a chromosome-specific chimpanzee alpha satellite subset: Evolutionary relationship to subsets on human chromosomes. Genomics 1996, 33, 220–228.
- Haaf, T.; Warburton, P.E.; Willard, H.F. Integration of human α-satellite DNA into simian chromosomes: Centromere protein binding and disruption of normal chromosome segregation. Cell 1992, 70, 681–696.Haaf, T.; Willard, H.F. Chromosome-specific α-satellite DNA from the centromere of chimpanzee chromosome 4. Chromosoma 1997, 106, 226–232.
- Warburton, P.E.; Haaf, T.; Gosden, J.; Lawson, D.; Willard, H.F. Characterization of a chromosome-specific chimpanzee alpha satellite subset: Evolutionary relationship to subsets on human chromosomes. Genomics 1996, 33, 220–228.Terada, S.; Hirai, Y.; Hirai, H.; Koga, A. Higher-order repeat structure in alpha satellite DNA is an attribute of hominoids rather than hominids. J. Hum. Genet. 2013, 58, 752–754.
- Haaf, T.; Willard, H.F. Chromosome-specific α-satellite DNA from the centromere of chimpanzee chromosome 4. Chromosoma 1997, 106, 226–232.Alkan, C.; Cardone, M.F.; Catacchio, C.R.; Antonacci, F.; O’Brien, S.J.; Ryder, O.A.; Purgato, S.; Zoli, M.; Della Valle, G.; Eichler, E.E.; et al. Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genom. Res. 2011, 21, 137–145.
- Terada, S.; Hirai, Y.; Hirai, H.; Koga, A. Higher-order repeat structure in alpha satellite DNA is an attribute of hominoids rather than hominids. J. Hum. Genet. 2013, 58, 752–754.Koga, A.; Tanabe, H.; Hirai, Y.; Imai, H.; Imamura, M.; Oishi, T.; Stanyon, R.; Hirai, H. Co-opted megasatellite DNA drives evolution of secondary night vision in Azara’s Owl monkey. Genome Biol. Evol. 2017, 9, 1963–1970.
- Alkan, C.; Cardone, M.F.; Catacchio, C.R.; Antonacci, F.; O’Brien, S.J.; Ryder, O.A.; Purgato, S.; Zoli, M.; Della Valle, G.; Eichler, E.E.; et al. Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genom. Res. 2011, 21, 137–145.Nishihara, H.; Stanyon, R.; Kusumi, J.; Hirai, H.; Koga, A. Evolutionary origin of OwlRep, a megasatellite DNA associated with adaptation of owl monkeys to nocturnal lifestyle. Genome Biol. Evol. 2018, 10, 157–165.
- Koga, A.; Tanabe, H.; Hirai, Y.; Imai, H.; Imamura, M.; Oishi, T.; Stanyon, R.; Hirai, H. Co-opted megasatellite DNA drives evolution of secondary night vision in Azara’s Owl monkey. Genome Biol. Evol. 2017, 9, 1963–1970.Cacheux, L.; Ponger, L.; Gerbault-Seureau, M.; Loll, F.; Gey, D.; Richard, F.A.; Escudé, C. The targeted sequencing of alpha satellite DNA in Cercopithecus pogonias provides new insight into the diversity and dynamics of centromeric repeats in old world monkeys. Genome Biol. Evol. 2018, 10, 1837–1851.
- Nishihara, H.; Stanyon, R.; Kusumi, J.; Hirai, H.; Koga, A. Evolutionary origin of OwlRep, a megasatellite DNA associated with adaptation of owl monkeys to nocturnal lifestyle. Genome Biol. Evol. 2018, 10, 157–165.Waye, J.S.; Willard, H.F. Human β satellite DNA: Genomic organization and sequence definition of a class of highly repetitive tandem DNA. Proc. Natl. Acad. Sci. USA 1989, 86, 6250–6254.
- Cacheux, L.; Ponger, L.; Gerbault-Seureau, M.; Loll, F.; Gey, D.; Richard, F.A.; Escudé, C. The targeted sequencing of alpha satellite DNA in Cercopithecus pogonias provides new insight into the diversity and dynamics of centromeric repeats in old world monkeys. Genome Biol. Evol. 2018, 10, 1837–1851.Greig, G.M.; Willard, H.F. β satellite DNA: Characterization and localization of two subfamilies from the distal and proximal short arms of the human acrocentric chromosomes. Genomics 1992, 12, 573–580.
- Waye, J.S.; Willard, H.F. Human β satellite DNA: Genomic organization and sequence definition of a class of highly repetitive tandem DNA. Proc. Natl. Acad. Sci. USA 1989, 86, 6250–6254.Cardone, M.F.; Ballarati, L.; Ventura, M.; Rocchi, M.; Marozzi, A.; Ginelli, E.; Meneveri, R. Evolution of beta satellite DNA sequences: Evidence for duplication-mediated repeat amplification and spreading. Mol. Biol. Evol. 2004, 21, 1792–1799.
- Greig, G.M.; Willard, H.F. β satellite DNA: Characterization and localization of two subfamilies from the distal and proximal short arms of the human acrocentric chromosomes. Genomics 1992, 12, 573–580.Meneveri, R.; Agresti, A.; Valle, G.D.; Talarico, D.; Siccardi, A.G.; Ginelli, E. Identification of a human clustered G + C-rich DNA family of repeats (Sau3A family). J. Mol. Biol. 1985, 186, 483–489.
- Cardone, M.F.; Ballarati, L.; Ventura, M.; Rocchi, M.; Marozzi, A.; Ginelli, E.; Meneveri, R. Evolution of beta satellite DNA sequences: Evidence for duplication-mediated repeat amplification and spreading. Mol. Biol. Evol. 2004, 21, 1792–1799.Meneveri, R.; Agresti, A.; Marozzi, A.; Saccone, S.; Rocchi, M.; Archidiacono, N.; Corneo, G.; Valle, G.D.; Ginelli, E. Molecular organization and chromosomal location of human GC-rich heterochromatic blocks. Gene 1993, 123, 227–234.
- Meneveri, R.; Agresti, A.; Valle, G.D.; Talarico, D.; Siccardi, A.G.; Ginelli, E. Identification of a human clustered G + C-rich DNA family of repeats (Sau3A family). J. Mol. Biol. 1985, 186, 483–489.Agresti, A.; Meneveri, R.; Siccardi, A.G.; Marozzi, A.; Corneo, G.; Gaudi, S.; Ginelli, E. Linkage in human heterochromatin between highly divergent Sau3A repeats and a new family of repeated DNA sequences (HaeIII family). J. Mol. Biol. 1989, 205, 625–631.
- Meneveri, R.; Agresti, A.; Marozzi, A.; Saccone, S.; Rocchi, M.; Archidiacono, N.; Corneo, G.; Valle, G.D.; Ginelli, E. Molecular organization and chromosomal location of human GC-rich heterochromatic blocks. Gene 1993, 123, 227–234.Bakker, E.; Wijmenga, C.; Vossen, R.H.A.M.; Padberg, G.W.; Hewitt, J.; van Der Wielen, M.; Rasmussen, K.; Frants, R.R. The FSHD-linked locus D4F104S1 (p13E-11) ON 4q35 has a homologue on 10qter. Muscle Nerve 1995, 18, S39–S44.
- Agresti, A.; Meneveri, R.; Siccardi, A.G.; Marozzi, A.; Corneo, G.; Gaudi, S.; Ginelli, E. Linkage in human heterochromatin between highly divergent Sau3A repeats and a new family of repeated DNA sequences (HaeIII family). J. Mol. Biol. 1989, 205, 625–631.Lemmers, R.J.F.L.; Wohlgemuth, M.; Frants, R.R.; Padberg, G.W.; Morava, E.; Van Der Maarel, S.M. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2004, 75, 1124–1130.
- Bakker, E.; Wijmenga, C.; Vossen, R.H.A.M.; Padberg, G.W.; Hewitt, J.; van Der Wielen, M.; Rasmussen, K.; Frants, R.R. The FSHD-linked locus D4F104S1 (p13E-11) ON 4q35 has a homologue on 10qter. Muscle Nerve 1995, 18, S39–S44.Clark, L.N.; Koehler, U.; Ward, D.C.; Wienberg, J.; Hewitt, J.E. Analysis of the organisation and localisation of the FSHD-associated tandem array in primates: Implications for the origin and evolution of the 3.3 kb repeat family. Chromosoma 1996, 105, 180–189.
- Lemmers, R.J.F.L.; Wohlgemuth, M.; Frants, R.R.; Padberg, G.W.; Morava, E.; Van Der Maarel, S.M. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2004, 75, 1124–1130.Winokur, S.T.; Bengtsson, U.; Vargas, J.C.; Wasmuth, J.J.; Altherr, M.R. The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum. Mol. Genet. 1996, 5, 1567–1575.
- Clark, L.N.; Koehler, U.; Ward, D.C.; Wienberg, J.; Hewitt, J.E. Analysis of the organisation and localisation of the FSHD-associated tandem array in primates: Implications for the origin and evolution of the 3.3 kb repeat family. Chromosoma 1996, 105, 180–189.Ballarati, L.; Piccini, I.; Carbone, L.; Archidiacono, N.; Rollier, A.; Marozzi, A.; Meneveri, R.; Ginelli, E. Human genome dispersal and evolution of 4q35 duplications and interspersed LSau repeats. Gene 2002, 296, 21–27.
- Winokur, S.T.; Bengtsson, U.; Vargas, J.C.; Wasmuth, J.J.; Altherr, M.R. The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum. Mol. Genet. 1996, 5, 1567–1575.Meneveri, R.; Agresti, A.; Rocchi, M.; Marozzi, A.; Ginellil, E. Analysis of GC-rich repetitive nucleotide sequences in great apes. J. Mol. Evol. 1995, 40, 405–412.
- Ballarati, L.; Piccini, I.; Carbone, L.; Archidiacono, N.; Rollier, A.; Marozzi, A.; Meneveri, R.; Ginelli, E. Human genome dispersal and evolution of 4q35 duplications and interspersed LSau repeats. Gene 2002, 296, 21–27.Hirai, H.; Taguchi, T.; Godwin, A.K. Genomic differentiation of 18S ribosomal DNA and β-satellite DNA in the hominoid and its evolutionary aspects. Chromosom. Res. 1999, 7, 531–540.
- Meneveri, R.; Agresti, A.; Rocchi, M.; Marozzi, A.; Ginellil, E. Analysis of GC-rich repetitive nucleotide sequences in great apes. J. Mol. Evol. 1995, 40, 405–412.McLaughlin, C.R.; Chadwick, B.P. Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. 2011, 12, R37.
- Hirai, H.; Taguchi, T.; Godwin, A.K. Genomic differentiation of 18S ribosomal DNA and β-satellite DNA in the hominoid and its evolutionary aspects. Chromosom. Res. 1999, 7, 531–540.Agresti, A.; Rainaldi, G.; Lobbiani, A.; Magnani, I.; Di Lernia, R.; Meneveri, R.; Siccardi, A.G.; Ginelli, E. Chromosomal location by in situ hybridization of the human Sau3A family of DNA repeats. Hum. Genet. 1987, 75, 326–332.
- McLaughlin, C.R.; Chadwick, B.P. Characterization of DXZ4 conservation in primates implies important functional roles for CTCF binding, array expression and tandem repeat organization on the X chromosome. Genome Biol. 2011, 12, R37.Mitchell, A.R.; Gosden, J.R.; Ryder, O.A. Satellite DNA relationships in man and the primates. Nucleic Acids Res. 1981, 9, 3235–3249.
- Agresti, A.; Rainaldi, G.; Lobbiani, A.; Magnani, I.; Di Lernia, R.; Meneveri, R.; Siccardi, A.G.; Ginelli, E. Chromosomal location by in situ hybridization of the human Sau3A family of DNA repeats. Hum. Genet. 1987, 75, 326–332.Fowler, J.C.S.; Burgoyne, L.A.; Baker, E.G.; Riugenbergs, M.L.; Callen, D.F. Human Satellite III DNA: Genomic location and sequence homogeneity of the TaqI-deficient polymorphic sequences. Chromosoma 1989, 98, 266–272.
- Mitchell, A.R.; Gosden, J.R.; Ryder, O.A. Satellite DNA relationships in man and the primates. Nucleic Acids Res. 1981, 9, 3235–3249.
- Fowler, J.C.S.; Burgoyne, L.A.; Baker, E.G.; Riugenbergs, M.L.; Callen, D.F. Human Satellite III DNA: Genomic location and sequence homogeneity of the TaqI-deficient polymorphic sequences. Chromosoma 1989, 98, 266–272.
- Hu, H.; Li, B.; Duan, S. The alteration of subtelomeric DNA methylation in aging-related diseases. Front. Genet. 2019, 10, doi:10.3389/fgene.2018.00697.
- Näslund, K.; Saetre, P.; Von Salomé, J.; Bergström, T.F.; Jareborg, N.; Jazin, E. Genome-wide prediction of human VNTRs. Genomics 2005, 85, 24–35, doi:10.1016/j.ygeno.2004.10.009.
- Vergnaud, G.; Denoeud, F. Minisatellites: Mutability and genome architecture. Genome Res. 2000, 10, 899–907, doi:10.1101/gr.10.7.899.
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)(n), present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626, doi:10.1073/pnas.85.18.6622.
- Riethman, H. Human subtelomeric copy number variations. Cytogenet. Genome Res. 2009, 123, 244–252, doi:10.1159/000184714.
- Cuadrado, A.; Jouve, N. Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6×-triticale. Chromosom. Res. 1994, 2, 331–338, doi:10.1007/BF01552727.
- Louis, E.J.; Vershinin, A.V. Chromosome ends: Different sequences may provide conserved functions. BioEssays 2005, 27, 685–697, doi:10.1002/bies.20259.
- Brown, W.R.A.; MacKinnon, P.J.; Villasanté, A.; Spurr, N.; Buckle, V.J.; Dobson, M.J. Structure and polymorphism of human telomere-associated DNA. Cell 1990, 63, 119–132, doi:10.1016/0092-8674(90)90293-N.
- Jurka, J.; Pethiyagoda, C. Simple repetitive DNA sequences from primates: Compilation and analysis. J. Mol. Evol. 1995, 40, 120–126, doi:10.1007/BF00167107.
- Araujo, N.P.; De Lima, L.G.; Dias, G.B.; Kuhn, G.C.S.; De Melo, A.L.; Yonenaga-Yassuda, Y.; Stanyon, R.; Svartman, M. Identification and characterization of a subtelomeric satellite DNA in Callitrichini monkeys. DNA Res. 2017, 24, 377–385, doi:10.1093/dnares/dsx010.
- Cechova, M.; Harris, R.S.; Tomaszkiewicz, M.; Arbeithuber, B.; Chiaromonte, F.; Makova, K.D. High Satellite Repeat Turnover in Great Apes Studied with Short- And Long-Read Technologies. Mol. Biol. Evol. 2019, 36, 2415–2431, doi:10.1093/molbev/msz156.
- Royle, N.J.; Baird, D.M.; Jeffreys, A.J. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat. Genet. 1994, 6, 52–56, doi:10.1038/ng0194-52.
- Koga, A.; Hirai, Y.; Hara, T.; Hirai, H. Repetitive sequences originating from the centromere constitute large-scale heterochromatin in the telomere region in the siamang, a small ape. Heredity (Edinb) 2012, 109, 180–187, doi:10.1038/hdy.2012.28.
- Ventura, M.; Catacchio, C.R.; Sajjadian, S.; Vives, L.; Sudmant, P.H.; Marques-Bonet, T.; Graves, T.A.; Wilson, R.K.; Eichler, E.E. The evolution of African great ape subtelomeric heterochromatin and the fusion of human chromosome 2. Genome Res. 2012, 22, 1036–1049, doi:10.1101/gr.136556.111.
- Ventura, M.; Antonacci, F.; Cardone, M.F.; Stanyon, R.; D’Addabbo, P.; Cellamare, A.; Sprague, L.J.; Eichler, E.E.; Archidiacono, N.; Rocchi, M. Evolutionary formation of new centromeres in macaque. Science 2007, 316, 243–246, doi:10.1126/science.1140615.
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29, doi:10.1038/nrm.2015.5.
- Fanning, T.G.; Seuánez, H.N.; Forman, L. Satellite DNA sequences in the New World primate Cebus apella (Platyrrhini, Primates). Chromosoma 1993, 102, 306–311, doi:10.1007/BF00661273.
- Nishibuchi, G.; Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosom. Res. 2017, 25, 77–87, doi:10.1007/s10577-016-9547-3.
- Riethman, H.; Ambrosini, A.; Paul, S. Human subtelomere structure and variation. Chromosom. Res. 2005, 13, 505–515, doi:10.1007/s10577-005-0998-1.
- Kehrer-Sawatzki, H.; Cooper, D.N. Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosom. Res. 2008, 16, 41–56, doi:10.1007/s10577-005-0998-1.
- Stankiewicz, P.; Park, S.S.; Inoue, K.; Lupski, J.R. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 2001, 11, 1205–1210.
- Carbone, L.; Alan Harris, R.; Gnerre, S.; Veeramah, K.R.; Lorente-Galdos, B.; Huddleston, J.; Meyer, T.J.; Herrero, J.; Roos, C.; Aken, B.; et al. Gibbon genome and the fast karyotype evolution of small apes. Nature 2014, 513, 195–201, doi:10.1038/nature13679.
- Koehler, U.; Bigoni, F.; Wienberg, J.; Stanyon, R. Genomic reorganization in the concolor gibbon (hylobates concolor) revealed by chromosome painting. Genomics 1995, 30, 287–292, doi:10.1006/geno.1995.9875.
- Wienberg, J.; Jauch, A.; Lüdecke, H.J.; Senger, G.; Horsthemke, B.; Claussen, U.; Cremer, T.; Arnold, N.; Lengauer, C. The origin of human chromosome 2 analyzed by comparative chromosome mapping with a DNA microlibrary. Chromosom. Res. 1994, 2, 405–410, doi:10.1007/BF01552800.
- Cheng, Z.; Ventura, M.; She, X.; Khaitovich, P.; Graves, T.; Osoegawa, K.; Church, D.; DeJong, P.; Wilson, R.K.; Pääbo, S.; et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 2005, 437, 88–93, doi:10.1038/nature04000.
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of tandemly repetitive DNA: Functional roles. Chromosom. Res. 2015, 23, 463–477, doi:10.1007/s10577-015-9494-4.
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153, doi:10.1073/pnas.1518008112.
