The MOB family proteins are constituted by highly conserved eukaryote kinase signal adaptors involved in the regulation of cell cycle progression, cell proliferation versus proliferation, morphogenesis, and cell differentiation and are often essential both for cell and organism survival.
- MOB,cell polarity,cytokinesis,Hippo pathway
1. Introduction
A MOB protein was first identified in 1998 by Luca & Winey [1], following a two-hybrid screen in budding yeast Saccharomyces cerevisiae that detected a monopolar spindle one (Mps1) binder protein. It was not until 2005 that Lai et al. (2005) [2] identified the first metazoan Mob gene, designated Mob as tumor suppressor (Mats), in Drosophila melanogaster. Since then, research efforts have identified Mob genes in further species expanding this family in the eukaryotic lineage. Mob genes are found in variable numbers with fungi and fly, typically possessing three to four genes, while humans have up to seven [3,4,5].
The MOB family proteins are constituted by highly conserved eukaryote kinase adaptors that are often essential both for cell and organism survival. MOB proteins were first characterized as regulators of ploidy maintenance, helping to ensure proper chromosome segregation before mitotic exit and allowing the transition from mitosis to cytokinesis [1]. This family is also necessary for maintaining cell polarity and morphology during cell division [6,7]. In multicellular organisms, MOBs have been mostly characterized as tumor suppressors and agents of morphogenesis, frequently by regulating GCKII STE20 and NDR kinases in the Hippo signaling pathway. Notably, the Hippo pathway interplays with various signaling pathways, like Wnt, mTOR, Notch, Hedghog, and the STRIPAK complex [8,9,10]. These pathways control a myriad of cellular processes and their activity is largely dependent on intercellular communication. In metazoans, the canonical Hippo signaling pathway comprises a core kinase cascade that includes Salvador/SAV1 and Mats/MOB1 as signal adaptors. When the Hippo pathway is activated, Hippo/MST1/2 kinase together with Salvador/SAV1 activate by phosphorylation Mats/MOB1 in a complex with the Warts/LATS1/2 kinase that in turn phosphorylates Yorkie/YAP/TAZ transcriptional co-activators, preventing its translocation into the nucleus and avoiding the transcriptional activation of target genes (Figure 1). Since YAP/TAZ induce the transcription of anti-apoptotic and proliferation-associated genes, such as BIRC5/surviving, BIRC2/cIAP1, and MCL1, Hippo signaling activation results in tumor suppression [11,12]. Alternative interactions between adaptor proteins, GCKII STE20, and NDR kinases result in non-canonical Hippo pathway signaling. MOB adaptor proteins, GCKII STE20 kinases, and NDR kinases are highly conserved protein families from yeast to metazoans. In multicellular eukaryotes, MOB proteins have been mostly characterized in the context of canonical Hippo signaling. However, these proteins also regulate cell biology processes in these species, namely mitotic exit, centrosome duplication and chromosome segregation [13,14,15]. Significantly, most core Hippo pathway components present homologous proteins in unicellular eukaryotes. In these organisms, MOB co-activators, GCKII STE20 kinases, and NDR kinases have been mainly characterized in the context of the MEN/SIN, both ensuring the correct genetic material distribution and cytokinesis (Figure 1). Interestingly, both MEN and SIN networks have a Cdc14 phosphatase as an effector protein, a highly conserved protein also present in multicellular eukaryotes [16].
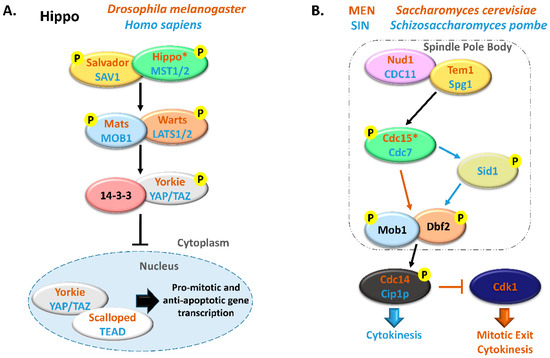
Figure 1. Comparison between Hippo and MEN/SIN pathways. (A) Schematic representation of the Hippo pathway in Drosophila melanogaster (orange) and in Homo sapiens (blue). (B) Schematic representation of the Mitotic Exit Network (MEN) in Saccharomyces cerevisiae (orange) and Septation Initiation Network (SIN) in Schizosaccharomyces pombe (blue). In these pathways, ortholog proteins are represented with oval nodes of the same color (yellow, Salvador/SAV1/Tem1/Spg1; green, Hippo/MST1/2/Cdc15/Cdc7; blue, Mats/MOB1/Mob1; orange, Warts/LATS1/2/Dbf2). * There are some controversies in the literature regarding whether to consider Cdc15 and Hippo as orthologous. This scheme is in accordance with Hergovich, 2017 [17].
Extensive reviews on MEN and Hippo signaling pathways have been published recently and we would like to direct the readers to them [5,18,19,20,21]. MEN/SIN and Hippo pathways have been mostly considered independent signal transduction cascades but the existence of several highly conserved proteins in these pathways suggests a shared functionality.
2. Functions of MOB Proteins
2.1. Tissue Homeostasis: MOB as Regulators of Cell Proliferation and Apoptosis
MOB proteins are tumor suppressors playing a critical role in regulating tissue homeostasis maintenance. In D. melanogaster, it was shown that DmMOB1 (Mats) controls cell proliferation and apoptosis through interaction with DmLATS (Warts), and its lethal depletion phenotype is rescued by HsMOB1 showing function conservation from invertebrates to vertebrates [2,26]. Indeed, H. sapiens, HsMOB1 also presents tumor suppressor activity, by phosphorylating HsLATS1, which can be triggered by HsMST1/2 phosphorylation but this is not essential (Figure 2 and Table 1) [27,28]. HsMOB1 tumor suppressor activity involves apoptotic signaling through Hippo pathway activation [29]. DmMOB1 tumor suppressor activity independent of DmMST (Hippo) phosphorylation also occurs in D. melanogaster [26]. Both HsMOB1 and HsMOB2 have been implicated in tissue growth suppression in cancer development [30,31,32,33]. Conversely, Chen et al. (2018) [34] showed that the cancer promoter complex HsMST4-HsMOB4/Phocein negatively regulates the tumor-suppressing complex HsMST1-HsMOB1 in pancreatic cancer. HsMOB4/Phocein and HsMST4 integrate the Striatin-interacting phosphatase and kinase (STRIPAK) complex [35]. The STRIPAK complex also includes the protein phosphatase PP2A and regulates vesicular trafficking, microtubule cytoskeleton and morphogenesis [9]. HsMOB3 also shows tumorigenic properties in glioblastoma cells by suppressing HsMST1 activity [36]. In Mus musculus, MmMOB1 functions as a tumor suppressor and tissue homeostasis factor as a member of Hippo signaling, namely by controlling apoptotic signaling in keratinocytes [10,37,38,39]. MmMOB1 also participates in renal homeostasis, MmMOB1 mediated Hippo activation, through MmLATS1 and MmYAP phosphorylation, is associated with diminished renal fibrosis [40]. The Wnt (wingless integrated) pathway is also activated but seems to have an opposite association. In Canis familiaris, CfMOB1-CfLATS1 Hippo signaling appears to regulate photoreceptor homeostasis [41]. In Gallus gallus, GgMOB2 interacts with GgSAV1 which acts as a growth suppressor through Hippo signaling [42]. Arabidopsis thaliana AtMOB1 regulates plant growth and development, and tissue homeostasis through interaction with AtMST, known as SIK1 [43,44,45,46]. In other angiosperms, Medicago sativa, MsMOB1 also regulates cell proliferation [47]. However, MsMob1 does not complement budding yeast MOB1 temperature sensitive growth phenotype. In the fungus Neurospora crassa, NcMob1 gene deletion results in overall reduced mycelium growth [3]. NcMob2 gene deletion exhibited phenotypes similar to NcMob1 but with less intensity. NcMob4 gene deletion resulted in a very mild decrease in tissue growth. Aspergillus nidulans AnMOB4/Phocein, which integrates the STRIPAK complex, also showed tumor suppressor properties [22]. Overall, several MOB isotypes are involved in the regulation of cell proliferation and apoptosis and its activation induces inhibition or promotion of tissue growth.
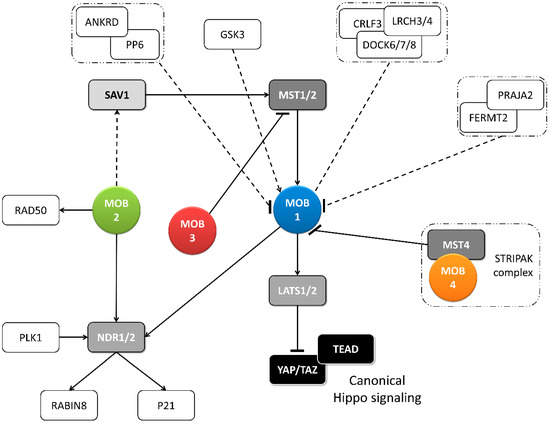
Figure 2. Metazoan MOB proteins present extensive regulation between isotypes. The canonical Hippo pathway is activated by upstream signals resulting in MST1/2 phosphorylation (which may be mediated by SAV1) and MOB1-LATS1/2 activation, causing YAP/TAZ phosphorylation and cytoplasm retention. The lack of YAP/TAZ transcriptional signaling results in a tumor suppressing effect. However, MOB proteins present several activities beyond canonical Hippo signaling. These include non-canonical Hippo signaling through different interactions with GCKII STE20 or NDR kinases, direct MOB stimulation by upstream signals and direct stimulation by MOBs of non-Hippo proteins. This intricate network results in direct and indirect MOB to MOB regulation. PLK1 regulates mitotic spindle orientation through NDR1 phosphorylation which results in NDR1 binding shifting from MOB1 to MOB2, favoring canonical Hippo activation [48]. NDR1/2 also regulate P21 and RABIN8 [49,50]. MOB3 is a MOB1 antagonist by inhibiting MST1 [36]. MOB4/Phocein-MST4, part of the STRIPAK complex, also antagonizes MOB1, by disrupting MOB1-MST1/1 binding [34]. MOB1 interacts in a HsMOB1-PPP6R1/2/3-ANKRD28 complex which appears to inhibit MOB1 mediated Hippo activation and in a DOCK6/7/8-CRLF3-LRCH3/4 complex, in a phosphorylation dependent manner [51,52]. Both PP6 phosphatase and DOCK6-8 promote actin cytoskeleton polarization signaling via RAC1. The FERMT2-PRAJA2 complex inhibits Hippo signaling by promoting MOB1 ubiquitin-proteasome degradation [40]. MOB1 is stimulated by GSK3β, a signaling hub involved in Wnt, mTOR, and Notch signaling [53]. MOB2 interacts with RAD50 stimulating the DNA damage response [54]. MOB2-SAV1 interaction was detected in G. gallus [42]. Notably, PP6, DOCK6, FERMT2 and GSK3 are proteins involved in cell-cell junctions. Colored boxes represent Hippo pathway members while other proteins are represented by non-colored boxes. Protein complexes are identified by an interrupted box with dashes and dots lines. Arrows represent activation while dashes represent inhibition. Lack of arrow or dash indicates an uncertain effect. Full lines represent well-established interactions. Interrupted lines represent less documented interactions.
Table 1. MOB functions in multicellular organisms.
| Protein | Described Functions | Functional Category | References | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TH | M | D | CC | ||||||||||||||||||||||||||
| Mammals and birds | HsMOB1 | Tissue growth (suppressor); apoptotic signaling; mitotic exit; protein complex positioning to spindle midzone, mitotic spindle orientation; centrosome duplication and disjunction; cytokinesis | X | X | [ | 13 | , | 14 | , | 27 | , | 28 | , | 29 | , | 30 | , | 34 | , | 48 | , | 55 | , | 56 | , | 57 | ] | ||
| HsMOB2 | Tissue growth (suppressor); cortical development; mitotic spindle orientation; centrosome duplication; DNA damage response | X | X | X | [ | 14 | , | 33 | , | 48 | , | 54 | , | 58 | , | 59 | , | 60 | , | 61 | ] | ||||||||
| HsMOB3 | Tissue growth (enhancer through MST1 inhibition); apoptotic signaling | X | X | [ | 14 | , | 36 | ] | |||||||||||||||||||||
| HsMOB4 | Tissue growth (enhancer through MST1-MOB1 complex inhibition) | X | [ | 34 | , | 35 | ] | ||||||||||||||||||||||
| MmMOB1 | Tissue growth (suppressor); lung and neuronal morphogenesis; stem cell differentiation and maintenance; centrosome duplication; actin cytoskeleton polarization | X | X | X | X | [ | 10 | , | 37 | , | 38 | , | 39 | , | 40 | , | 53 | , | 62 | , | 63 | ] | |||||||
| MmMOB2 | Neuronal and cortical development; actin cytoskeleton organization. | X | [ | 59 | , | 64 | ] | ||||||||||||||||||||||
| MmMOB4 | Dendritic arborization in neuronal development | X | [ | 34 | ] | ||||||||||||||||||||||||
| CfMOB1 | Photoreceptor growth; mitosis | X | X | [ | 41 | ] | |||||||||||||||||||||||
| GgMOB2 | Tissue growth (suppressor); follicle development | X | X | [ | 42 | ] | |||||||||||||||||||||||
| Insects | DmMOB1 | Tissue growth (suppressor); stem cell differentiation; chromosome segregation | X | X | X | [ | 2 | , | 15 | , | 26 | , | 62 | , | 65 | ] | |||||||||||||
| DmMOB2 | Wing hair, photoreceptor and neuromuscular junction morphogenesis | X | [ | 66 | , | 67 | , | 68 | ] | ||||||||||||||||||||
| DmMOB4 | Synapse morphogenesis and microtubule organization; spindle pole assembly; axonal transport of autophagosomes | X | X | [ | 69 | , | 70 | , | 71 | ] | |||||||||||||||||||
| Plants | AtMOB1 | Plant growth (enhancer); development of stem and root; sporogenesis and gametogenesis; stem cell differentiation and maintenance; apoptotic signaling; cytokinesis | X | X | X | X | [ | 43 | , | 44 | , | 45 | , | 46 | ] | ||||||||||||||
| MsMOB1 | Tissue growth (suppressor); stem cell association | X | X | [ | 47 | , | 72 | ] | |||||||||||||||||||||
| Fungi | NcMOB1 | Tissue growth (enhancer); septum, aerial mycelium and fruiting body development; conidiation; ascosporogenesis; meiosis | X | X | X | X | [ | 3 | ] | ||||||||||||||||||||
| NcMOB2 | Tissue growth (enhancer); hypha development; conidiation | X | X | X | [ | 3 | ] | ||||||||||||||||||||||
| NcMOB4 | Tissue growth (enhancer); fruiting body development; vegetative cell fusion | X | X | [ | 3 | , | 24 | ] | |||||||||||||||||||||
| AnMOB4 | Tissue growth (suppressor); ascosporogenesis; meiosis. | X | X | X | [ | 22 | ] | ||||||||||||||||||||||
| SmMOB4 | Vegetative cell fusion; conidiation and ascosporogenesis; meiosis | X | X | X | [ | 23 | ] | ||||||||||||||||||||||
| ChMOB2 | Conidiation and ascosporogenesis; meiosis | X | X | [ | 4 | ] | |||||||||||||||||||||||
TH—tissue homeostasis, M—morphogenesis, D—differentiation, CC—cell cycle progression, Dm—Drosophila melanogaster, Hs—Homo sapiens, Mm—Mus musculus, Cf—Canis familiaris, Gg—Gallus gallus, At—Arabidopsis thaliana, Ms—Medicago sativa, Nc—Neurospora crassa, An—Aspergillus nidulans, Sm—Sordaria macrospora, Ch—Colletotrichum higginsianum.
2.2. Morphogenesis: A MOB Function in Various Species and Cell Types
MOB proteins are also important cell polarity and tissue morphogenesis regulators (Table 1). D. melanogaster DmMOB2 interacts with DmNDR (Trc) and is necessary for wing hair, photoreceptor and neuromuscular junction morphogenesis [66,67,68]. DmMOB4/Phocein is also necessary for synapse morphogenesis and microtubule organization [69]. In H. sapiens, HsMOB2 loss of function mutations are associated with defective cortical development [59]. Interestingly, a similar phenotype is produced by mutations in cell adhesion protocadherins FAT4-DCHS1 which is compensated by Yap knockdown [73]. M. musculus MmMOB1 controls lung morphogenesis through YAP/TAZ regulation and neuritogenesis independent of YAP [53,62]. MmMOB1 mediated neuritogenesis is stimulated by GSK3β (glycogen synthase kinase3β) [53]. Notably, GSK3 functions as a signaling hub, integrated into Wnt, mTOR (mammalian target of rapamycin, and Notch pathways [74]. MmMOB2 is necessary for neuritogenesis and cortical development, including ciliogenesis [59,64]. MmMOB2 RNAi and overexpression studies showed this protein’s role in neuritogenesis is synergistic with MmNDR2 and affects the actin cytoskeleton [64]. Contrary to H. sapiens, M. musculus MmDCHS1 mutation phenotype could not be compensated by YAP knockdown [59]. MmMOB4/Phocein regulates dendritic arborization in neuronal development through STRIPAK complex signaling [25]. In G. gallus, GgSAV1-GgMOB2 interaction also regulates follicle development [42]. A. thaliana, AtMOB1 is necessary for plant structural development of stem and root [43,44,46]. Also, in fungi, MOB proteins participate in morphogenesis. In N. crassa NcMOB1 is necessary for septum and aerial mycelium formation and conidiation with NcMob1 gene deletion resulting in increased hyphae branching despite decreased mycelium growth. NcMOB2 controls hyphae polar tip extension through regulation of the NDR kinase NcCOT1 [3]. NcMOB4/Phocein is essential for vegetative cell fusion and consequent fruiting body formation in a way unrelated to NDR signaling. NcMOB1 and NcMOB4/Phocein both participate in fruiting body morphogenesis but NcMOB1 activity occurs by interaction with NcLATS (DBF2) while NcMOB4/Phocein integrates the STRIPAK complex [3,24]. Moreover, NcMOB4/Phocein and SmMOB4/Phocein of the Ascomycete Sordaria macrospora also control vegetative cell fusion [3,23]. These data show that the involvement of MOB proteins in morphogenesis is not exclusive of a specific MOB isotype, but that different MOB isotypes participate in this process (Table 1).
2.3. Cell Cycle Progression: MOBs as Regulators of Mitosis, Cytokinesis, and Centrosome Biology
Several core components of the Hippo pathway have been implicated in the regulation of eukaryotic cell cycle progression, including MOB proteins (Figure 2 and Figure 3) [17]. DmMOB1 null mutants present aberrant chromosome segregation during embryogenesis [15] while DmMOB4/Phocein depleted cells fail to properly assemble the spindle pole [70]. DmMOB4/Phocein also integrates the STRIPAK complex and is necessary for PP2A regulated axonal transport of autophagosomes [71]. DmMOB1 localizes to the cytoplasm and nucleus, but also to the centrosome [15]. HsMOB1 is required for mitotic exit and its depletion results in prolonged telophase [13]. The same is true for its binding partner HsLATS1/2. HsMOB1 is necessary for correct positioning of the mitotic regulator chromosomal passenger complex to the spindle midzone during anaphase [56]. Correct mitotic spindle orientation is regulated through NDR1 phosphorylation by PLK1 which results in NDR1 binding shifting from HsMOB1 to HsMOB2 [48]. HsMOB1/HsMOB2 competitive binding to NDR1 was also observed in centrosome duplication [14]. HsMOB2 binding to NDR seems to inhibit its activation which occurs through phosphorylation [60]. HsMOB1 has also been implicated in centrosome disjunction by interfering with NEK2 centrosome localization [57]. HsMOB1 also regulates correct cell abscission and cytokinesis [55]. HsMOB1 knockdown cells show increased motility immediately after telophase/cytokinesis and persist connected by long intercellular bridges [55]. Also, MOB1 depletion results in centriole separation which supports the idea that MOB1 is required for centriole rejoining at the end of mitosis. Interestingly, components of the Hippo pathway, HsSAV1 and MST2 cooperate with the NEK2 kinase to regulate centrosome disjunction [57]. Additionally, HsMOB1 interacts with serine/threonine phosphatases PP6 and an ankyrin repeat–containing protein ANKRD, forming a HsMOB1-PPP6R1/2/3-ANKRD28 complex. Also, interactions with leucine-rich repeats and calponin homology domain–containing proteins LRCH, cytokine receptor–like factor 3 and dedicator of cytokinesis proteins DOCK forming a HsDOCK6/7/8-CRLF3-LRCH3/4 complex were described [51,52]. Time point analysis suggests the PP6 complex may inhibit HsMOB1 mediated Hippo activation. HsMOB1 is mostly localized in the cytoplasm and to a lesser extent in the cytoplasmic membrane, but it also localizes to various cell cycle associated structures, namely centrosome, kinetochores in early mitosis and spindle midzone in late mitosis [13,14,56,61]. HsMOB2 induces G1/S cell cycle arrest in response to DNA damage, independently of NDR kinases [54]. Coincidently, HsMOB2 localizes to the nucleus [14,61]. HsMOB3, a cytoplasmic protein, prevents high cell density growth inhibition by downregulating HsMST1 mediated apoptotic signaling in glioblastoma cells [14,36]. Similarly to human, in mouse MmMOB1 regulates centrosome duplication in keratinocytes, through the regulation of MmLATS and MmYAP [37]. MmMOB1 also interacts with MmDOCK8 in thymocytes, stimulating MmRAC1 actin cytoskeleton polarization signaling [63]. MmMOB1-MmMST1/2 mediate MmRhoA GTP charging and MmRAC1 signaling stimulation. C. familiaris CfMOB1 is also necessary for correct mitosis in photoreceptor cells [41]. In plants A. thaliana and M. sativa, AtMOB1 and MsMOB1 participate in apoptotic signaling during meiotic microsporogenesis and macrosporogenesis and also regulate cytokinesis [43,46,47]. Supporting its role in cytokinesis MsMOB1 is cytoplasmic, but localizes to the cell plate during septum formation and at spindle microtubule structures related to cytokinesis [47]. AtMOB1 is present at the cytoplasm, cytoplasmic membrane, and nucleus [43,44]. A. thaliana Hippo/MST1/2 homolog SIK1 complements S. cerevisiae Ste20 deletion in mitotic exit and SIK1 was shown to bind to AtMOB1 [45]. In conclusion, MOB proteins play critical roles in accurate cell division and cytokinesis and, besides other cellular localizations, tend to localize at the centrosome when this structure is present, a localization that is shared by other Hippo components (Table 1).
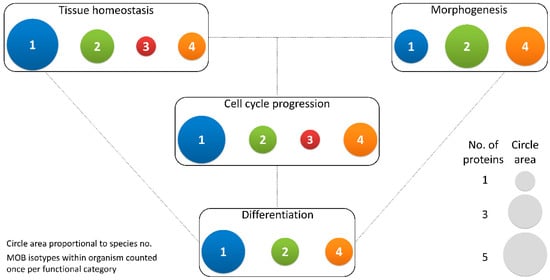
Figure 3. Different MOB isotypes present similar functions. MOB functions were divided into 4 categories: tissue homeostasis, morphogenesis, differentiation and cell cycle progression (includes mitosis, meiosis, cytokinesis, and centrosome biology). Circle areas are proportional to the number of proteins counted in the literature related to these functions, as is illustrated in the protein count examples at the bottom right. MOB protein isotypes for each organism were counted only once per function category (variants within protein isotypes were not considered). It is obvious that there is no one MOB isotype responsible for a particular function. Various MOB isotypes present similar functions, be it in distinct species or in distinct tissues in the same species. In fungi, NcMOB1, ChMOB2, and SmMOB4/Phocein, all participate in conidiation, ascosporogenesis, and meiosis [3,4,23]. In M. musculus, MmMOB1, MmMOB2, and MmMOB4/Phocein are necessary for neuronal development while MmMOB1 also promotes lung morphogenesis [25,53,62,64]. The data also evidence a higher abundance of studies on MOB1 proteins, which are the more represented in each category, except morphogenesis. Colors representing MOB isotypes: 1—blue, 2—green, 3—red, 4—orange.
2.4. Differentiation and Stem Cell Maintenance: MOB in a Crossroad between Morphogenesis, Tissue Homeostasis, and Cell Cycle Regulation
In Drosophila DmMOB1 is essential for embryonic development [15]. This becomes evident after both zygotic and maternal DmMOB1 depletion that results in non-viable embryos indicating a probable role for DmMOB1 in stem cell differentiation. In mouse, MmMOB1 is necessary for embryonic stem cell differentiation into the three germ layers, an activity that is dependent on YAP regulation [65]. MmMOB1 is also involved in bronchioalveolar cell differentiation and alveolar stem cell maintenance through YAP/TAZ regulation [62]. Surprisingly, this last study did not find evidence that MmMST or MmLATS kinases were also involved in YAP/TAZ regulation. The plant AtMOB1 is necessary for sexual development, namely during sporogenesis and gametogenesis [43,44]. AtMOB1 depleted cells present reduced meristem length and cell number, suggesting a role not only in cell differentiation but also in stem cell maintenance, as documented in mouse [43,44,46]. AtMOB1 depleted cells also present a substantially reduced seed output [43]. The protein localizes to meiocytes, supporting its participation in meiosis. Interestingly, in M. sativa MsMOB1 localizes to the meristem in a cell cycle dependent manner [47]. Fungi MOB proteins are also involved in the differentiation of cells for sexual development and the meiotic process. The NcMOB1, Colletotrichum higginsianum ChMOB2 and SmMOB4/Phocein are involved in conidiation, ascosporogenesis, and meiosis [3,4,23]. NcMOB2 exhibited similar properties as NcMOB1 in conidiation, but not in ascosporogenesis and meiosis [3]. ChMOB2 interacts with ChNDR, known as Cbk1 [4]. A. nidulans AnMOB4/Phocein is also necessary for ascospore production through meiosis [22].
