Stilbenes are a small family of polyphenolic secondary metabolites that can be found in several distantly related plant species. These compounds act as phytoalexins, playing a crucial role in plant defense against phytopathogens, as well as being involved in the adaptation of plants to abiotic environmental factors. Among stilbenes, trans-resveratrol is certainly the most popular and extensively studied for its health properties. However, many other stilbenes, both monomeric and oligomeric, are currently under intensive investigation due to their biological role and bioactivity.
- stilbenes
- secondary metabolites
- polyphenols
- phytoalexins
- phenylpropanoid pathway
- stilbene biosynthesis
- stilbene synthase
- resveratrol synthase
- pinosylvin synthase
Note:All the information in this draft can be edited by authors. And the entry will be online only after authors edit and submit it.
1. Introduction
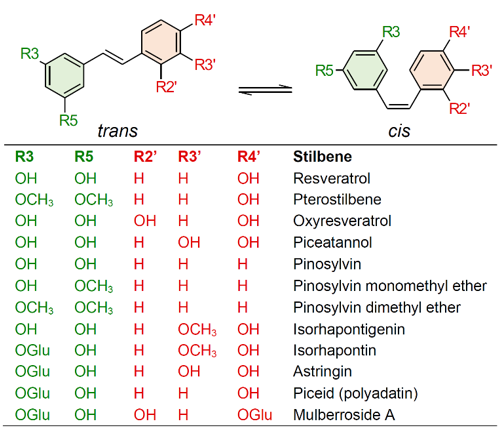
Stilbenes are a small yet important class of non-flavonoid polyphenols, sharing a common structure characterized by a 14-carbon skeleton composed of two benzene rings linked by an ethylene bridge (Figure 1). Due to the presence of the central ethylene moiety between the aromatic rings, stilbenes exist as the two possible stereoisomers cis and trans. However, the naturally occurring stilbenes are usually in the trans form [1]. Plant stilbenes, together with other polyphenols such as flavonoids, isoflavonoids, curcuminoids, and xanthones, belong to the class of polyketides. Over 400 different stilbene compounds are currently known [2], mostly derived from trans-resveratrol (3,5,4′-trihydroxy-trans-stilbene) (Figure 1), although different structures can be found in specific plant families [3].
Stilbenes have been identified in at least 72 plant species belonging to 31 genera and 12 distantly related families, including Pinaceae (e.g., Picea abies (L.) Karst. and Pinus nigra J.F. Arnold), Gnetaceae (e.g., Gnetum parvifolium (Warb.) W.C. Cheng and G. africanum Welw.), Fabaceae (Arachis hypogaea L. and Robinia pseudoacacia L.), Vitaceae (e.g., Vitis vinifera L. and V. amurensis Rupr.), Moraceae (e.g., Morus alba L. and M. macroura Miq.), and Polygonaceae (e.g., Polygonum cuspidatum Sieb. et Zucc. and P. multiflorum Thunb.) [4][5][4,5]. Given their nutraceutical value, stilbene content and composition have mainly been investigated in food plants, and the knowledge of stilbene distribution in nature is still poor. This is partially related to the complexity of the quali-quantitative analysis of stilbenes, which is in turn related to the unavailability of standards and the detection limits of analytical methods [2]. For these reasons, most of the studies carried out to date have been focused on simple stilbenes, such as resveratrol, piceid, pterostilbene, and piceatannol (Figure 1). Current knowledge on the distribution of stilbenes in the plant kingdom will not be presented in this review, as this topic is covered by excellent recent reviews [4][5][4,5].
Figure 1.
Chemical structures of common stilbene monomer derivatives. (OGlu) O-β-D-glucopyranoside.
Stilbenes are mainly involved in constitutive and inducible protection of the plant against biotic (phytopathogenic microorganisms and herbivores) and abiotic (e.g., UV radiation and tropospheric ozone) stress [3][6][3,6]. On one side they counteract the aggression exerting a direct toxic effect on the pathogen, while on the other they act as antioxidants, protecting the cells from oxidative damage [7][8][9][7–9]. Stilbenes possess several antipathogenic properties including antibacterial, antifungal [10][11][10,11], nematocidal [12], and insecticidal [13][14][13,14]. They could also act as a deterrent towards vertebrate herbivory [15], as a possible negative effect of stilbenes has been reported on snowshoe hares (Lepus americanus Erxleben) [16][17][16,17] and field voles (Microtus agrestis L.) [18]. The role of stilbenes, among other polyphenols, in counteracting oxidative stress is just as important, as the plant response to pathogen attack involves the production of reactive oxygen species (ROS), which both act as signals for the activation of stress and defense pathways and as toxic substances capable of directly damaging the pathogen. Oxidative stress may also be induced by many abiotic conditions, such as drought, thermal stress, ultraviolet radiation, mechanical stress, heavy metals, salts, and air pollutants such as ozone [19]. Unsurprisingly, many of these factors also affect stilbene production [20].
Over the past 20 years, the bioactivities of stilbenes have been intensively investigated due to their impact on human health. Among stilbenes, resveratrol is the best known and the most studied. Basic scientific research and over 240 clinical studies have demonstrated the multiplicity of trans-resveratrol pharmacological effects, including antioxidant [21], anti-inflammatory [22], anticancer [23][24][23,24], estrogenic [25], neuroprotective [26], cardioprotective [27], anti-atherosclerotic [28], anti-aging [29], anti-diabetic [30], anti-osteoporosis [25], and anti-obesity properties [31]. In recent years, considerable attention has also been paid to other monomeric stilbenes, including pterostilbene [32], pinosylvin [33], and piceatannol [34], as well as to oligomeric stilbenes such as viniferins [35][36][35,36], which have been shown to possess similar and often more pronounced health-promoting properties than resveratrol.
Due to their potential use in the nutraceutical, cosmeceutical, and pharmaceutical fields, great interest is directed at the methods for large-scale production of stilbenes. For instance, it has been estimated that the global market for trans-resveratrol will almost double in the next 6 years, from 58 million USD in 2020 to 99.4 million USD by 2026 [37]. Methods for obtaining stilbenes can be grouped into three categories: direct extraction from plants, chemical synthesis, and the use of biotechnologies. The chemical synthesis of stilbenes has been reported, but this method is not economically feasible, in addition to being difficult in terms of stereospecific synthesis [38][39][38,39]. Considerable efforts have been devoted to the development of biotechnological methods for stilbene production, which broadly include tissue culture techniques [40], biotransformation [41], and metabolic engineering [42]. Nevertheless, the major way of supplying stilbenes is the direct extraction from plants such as P. cuspidatum and V. vinifera [43].
2. Biosynthesis of Stilbenes and Stilbenoids
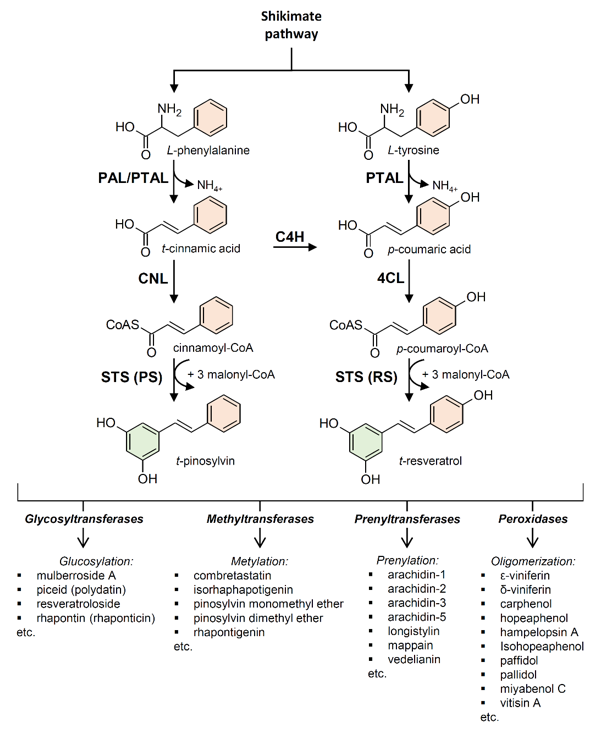
Stilbenes and stilbenoids are biosynthesized through the phenylpropanoid pathway, which is also responsible for the biosynthesis of numerous primary and secondary metabolites including flavonoids, coumarins, hydrolyzable tannins, monolignols, lignans, and lignins [44]. Generated by the shikimate pathway, the aromatic amino acid L-phenylalanine is the primary starting molecule of the phenylpropanoid pathway (Figure 2). The non-oxidative deamination of L-phenylalanine to form trans-cinnamic acid, catalyzed by phenylalanine ammonia-lyase (PAL; EC 4.3.1.24), is the entry step for the carbon channeling from primary metabolism into the phenylpropanoid secondary metabolism. PAL is ubiquitous in plants [45], and it is undoubtedly the most studied enzyme involved in plant secondary metabolism [46]. Cinnamic acid can be bound to a coenzyme A (CoA) molecule by cinnamate:CoA ligase (CNL; EC 6.2.1.-) to form cinnamoyl-CoA. Alternatively, cinnamic acid can be hydroxylated by cinnamate 4‐hydroxylase (C4H), a cytochrome P450 enzyme (EC 1.14.14.91), to form p-coumaric acid. Some plants (mainly monocots but also dicots) possess a bifunctional phenylalanine/tyrosine ammonia-lyase (PTAL, EC 4.3.1.25) that efficiently deaminates both L-phenylalanine (PAL activity) and L-tyrosine (TAL activity) [47][48][49][50][47–50]. These plants can directly produce p-coumaric acid using L-tyrosine as a substrate, bypassing the requirement for L-phenylalanine and C4H. A molecule of CoA is then bound to p-coumaric acid by 4-coumarate: CoA ligase (4CL; EC 6.2.1.12), generating p-coumaroyl-CoA, which provides an active intermediate in numerous branches of the general phenylpropanoid pathway [51].
Figure 2. Stilbene biosynthesis in plants. (PAL) phenylalanine ammonia-lyase; (PTAL) bifunctional L-phenylalanine/L-tyrosine ammonia-lyase; (C4H) cinnamate 4‐hydroxylase; (4CL) 4-coumarate:CoA ligase; (CNL) cinnamate:CoA ligase; (STS) stilbene synthase; (RS) resveratrol synthase; (PS) pinosylvin synthase.
2.1. Stilbene Synthase
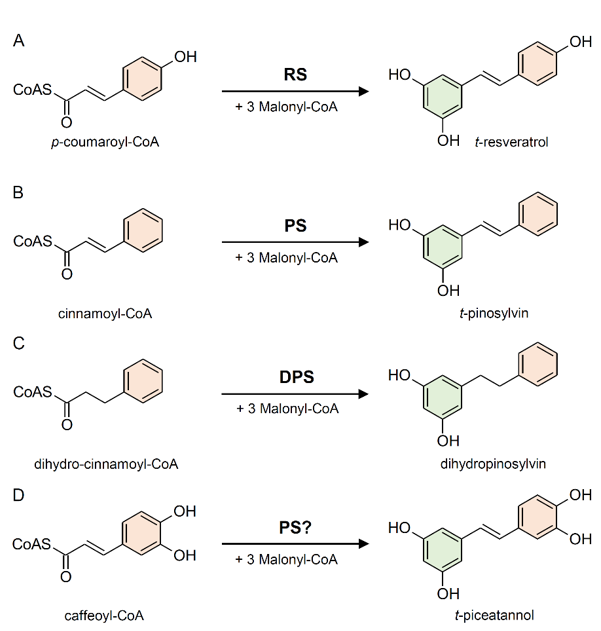
The enzyme stilbene synthases (STS) catalyze the direct formation of the stilbene skeleton through a single reaction from three units of malonyl-CoA and one CoA-ester of a cinnamic acid derivative (p-coumaroyl-CoA to form trans-resveratrol or cinnamoyl-CoA to form trans-pinosylvin) [52] (Figures 2 and 3). Malonyl-CoA is generated through a carboxylation reaction between acetyl-CoA and a bicarbonate ion (HCO3−) catalyzed by acetyl-CoA carboxylase (EC 6.4.1.2) in the presence of ATP (Figure 4).
Figure 3. Examples of reactions catalyzed by stilbene synthase enzymes. (A) Conversion of p-coumaroyl-CoA into t-resveratrol by resveratrol (RS) synthase (or trihydroxystilbene synthase I). (B) Conversion of cinnamoyl-CoA into t-pinosylvin by pinosylvin synthase (PS). (C) Conversion of dihydro-cinnamoyl-CoA into dihydropinosylvin by dihydro-pinosylvin synthase (DPS). (D) Conversion of caffeoyl-CoA into t-piceatannol, probably catalyzed by PS.
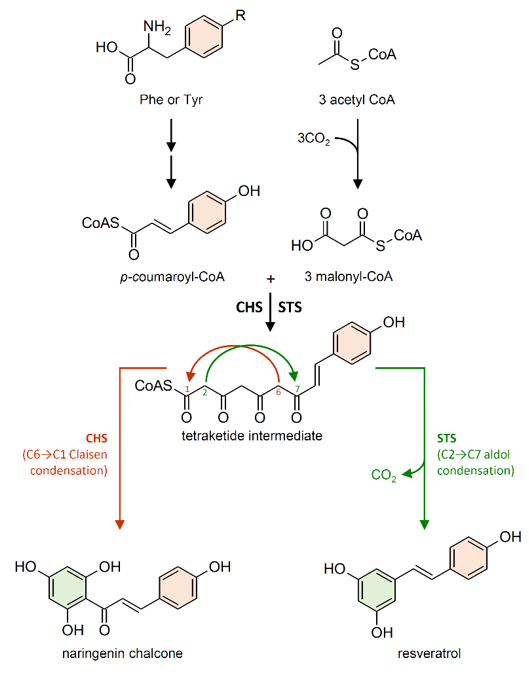
Figure 4. Reactions catalyzed by chalcone synthase (CHS) and stilbene synthase (STS) to produce naringenin chalcone and resveratrol, respectively. R = H phenylalanine (Phe); R = OH tyrosine (Tyr). Double arrows indicate multiple steps in the biosynthetic pathway.
Based on the preferred starting substrate, STS enzymes are classified into either a p-coumaroyl-CoA-specific type, such as trihydroxystilbene synthase I (also known as resveratrol synthase, EC 2.3.1.95), or a cinnamoyl-CoA-specific type, such as pinosylvin synthase (EC 2.3.1.146) (Figures 2 and 3). The former type has been mainly found in angiosperms like peanut [53], grapevine [54], and Tatar rhubarb (Rheum tataricum L.f) [55], while the latter type is typical in conifers and has been identified in several Pinus species like Scots pine (P. sylvestris L.) [56], Japanese red pine (P. densiflora Siebold & Zucc.) [57], and Eastern white pine (P. strobus L.) [58].
Pinus species can biosynthesize two types of stilbenes, i.e., pinosylvin and dihydropinosylvin, which are biosynthetically derived from cinnamoyl-CoA and dihydrocinnamoyl-CoA, respectively (Figure 3B,C). STS from P. strobus shows a clear preference for cinnamoyl-CoA and was therefore characterized as pinosylvin synthase [58]. Otherwise, STS from P. sylvestris shows an unusual preference for dihydro-cinnamoyl-CoA, identifying it as a dihydro-pinosylvin synthase [56]. STS does not exhibit absolute substrate specificity. While showing a preference for a given substrate, the same STS enzyme can accept different cinnamic acid derivatives as starting substrates catalyzing the biosynthesis of different stilbenes. For example, the enzyme responsible for the biosynthesis of piceatannol (3,5,3′,4′-tetrahydroxystilbene) has not been identified yet, however, pinosylvin synthase from P. strobus proved to be active with caffeoyl-CoA in vitro (Figure 3D), suggesting that it could be responsible for piceatannol biosynthesis in planta [58].
STS enzymes belong to the type III polyketide synthase superfamily (PKSs), which also includes chalcone synthase (CHS; EC 2.3.1.74) [59]. STS and CHS share a high degree of similarity both in their amino acid sequence identity (which reaches 75–90% depending on the species) and in their crystallographic structures [51][60][51,60]. CHS genes are present in the genome of all plants analyzed so far, while STS have been identified in a limited number of plant species, often phylogenetically unrelated. Converging lines of evidence indicate that CHS is the archetypal enzyme from which STS evolved multiple times independently in stilbene-producing plants, through gene duplication followed by functional divergence [60][61][62][60–62]. CHS and STS are the most investigated enzymes among PKSs and, due to their high sequence similarity, they are often referred to as the CHS/STS family [63][64][63,64].
Although it employs the same substrates as STS, CHS is responsible for the first committed step in the biosynthesis of flavonoid-type compounds. Both enzymes generate the same linear tetraketide intermediate. However, CHS catalyzes a C6→C1 Claisen condensation of the intermediate to produce naringenin chalcone, while STS catalyzes an alternative C2→C7 aldol condensation of the intermediate to form a stilbene backbone (Figure 4) [59][65][66][59,65,66].
STS was first extracted and purified from suspension cultures of peanut cells elicited with UV radiation [53]. Cloning of two peanut STS genes revealed a high sequence identity with CHS throughout the coding region and the presence of an intron at the same position as a conserved intron in CHS [67]. STS genes and cDNAs were subsequently cloned and characterized from grapevine cell suspension cultures [68] and Scots pine plantlets [56], both induced by fungal elicitors. At present, STS genes have been cloned from several plant species including mulberry (Morus notabilis C.K. Schneid and M. atropurpurea Roxb.) [42][66][42,66], Scots pine [69], white spruce (Picea glauca (Moench) Voss) [70], Norway spruce (Picea abies (L.) H. Karst.) [71], Japanese red pine [57], and sorghum (Sorghum bicolor (L.) Moench) [72]. To the best of our knowledge, sorghum is the only monocot plant in which an STS gene (SbSTS1) has been identified.
In most stilbene-producing plants, STS exists as a small family consisting of 1–10 closely related paralogs. For example, the STS multigene family is represented by two members in white spruce [70] and Norway spruce [71], three members in Japanese red pine [57], almost five members in Scots pine [69], six members in peanut [73], and ten members in mulberry [66]. Remarkable exceptions to this role are sorghum, in whose genome only one STS gene has been identified [74][75][74,75], and grapevine, which possesses an uncommonly large number of STS genes. Both grapevine and sorghum genomes have been entirely sequenced [74][76][74,76]. Early Southern-blot analysis suggested that the grapevine STS gene family consisted of 15–20 members [77]. Genome-wide analysis carried out on the V. vinifera PN40024 genome led to the identification of 48 putative STS genes, designated VvSTS1 to VvSTS48, with at least 33 potentially coding for functional STS proteins [60][62][60,62].
To date, there is no evidence regarding the different substrate specificity and enzymatic activity of different VvSTSs. Functional characterization of nine VvSTSs confirmed that they encode for functional STS enzymes [62]. Since these nine genes were specifically chosen to represent the diversity of the VvSTS gene family, it is most likely that all grapevine VvSTSs encode enzymes with similar activity and specificity. Despite the high similarity between STS genes which makes it difficult to accurately distinguish the individual transcripts, gene expression studies revealed different transcriptional responses of distinct VvSTSs during development and in response to environmental stresses [60][78][79][60,78,79]. The expression of some VvSTSs was also found to be tissue-specific [60][79][60,79]. It is therefore likely that the large quantity of members in the grapevine STS gene family has evolved to allow for fine spatial and temporal regulation of stilbene biosynthesis under both normal and stress conditions.
References
- Roupe, K.A.; Remsberg, C.M.; Yáñez, J.A.; Davies, N.M. Pharmacometrics of stilbenes: Seguing towards the clinic. Clin. Pharmacol. 2006, 1, 81–101.
- El Khawand, T.; Courtois, A.; Valls, J.; Richard, T.; Krisa, S. A review of dietary stilbenes: Sources and bioavailability. Rev. 2018, 17, 1007–1029.
- Chong, J.; Poutaraud, A.; Hugueney, P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009, 177, 143–155.
- Rivière, C.; Pawlus, A.D.; Merillon, J.M. Natural stilbenoids: Distribution in the plant kingdom and chemotaxonomic interest in Vitaceae. Prod. Rep. 2012, 29, 1317–1333.
- Dubrovina, A.S.; Kiselev, K.V. Regulation of stilbene biosynthesis in plants. Planta 2017, 246, 597–623.
- Jeandet, P.; Delaunois, B.; Conreux, A.; Donnez, D.; Nuzzo, V.; Cordelier, S.; Clément, C.; Courot, E. Biosynthesis, metabolism, molecular engineering, and biological functions of stilbene phytoalexins in plants. Biofactors 2010, 36, 331–341.
- Waffo Teguo, P.; Fauconneau, B.; Deffieux, G.; Huguet, F.; Vercauteren, J.; Mérillon, J.M. Isolation, identification, and antioxidant activity of three stilbene glucosides newly extracted from Vitis vinifera cell cultures. Nat. Prod. 1998, 61, 655–657.
- Privat, C.; Telo, J.P.; Bernardes-Genisson, V.; Vieira, A.; Souchard, J.P.; Nepveu, F. Antioxidant properties of trans-ε-viniferin as compared to stilbene derivatives in aqueous and nonaqueous media. Agric. Food Chem. 2002, 50, 1213–1217.
- Biais, B.; Krisa, S.; Cluzet, S.; Da Costa, G.; Waffo-Teguo, P.; Mérillon, J.M.; Richard, T. Antioxidant and cytoprotective activities of grapevine stilbenes. Agric. Food Chem. 2017, 65, 4952–4960.
- Albert, S.; Horbach, R.; Deising, H.B.; Siewert, B.; Csuk, R. Synthesis and antimicrobial activity of (E) stilbene derivatives. Med. Chem. 2011, 19, 5155–5166.
- Chalal, M.; Klinguer, A.; Echairi, A.; Meunier, P.; Vervandier-Fasseur, D.; Adrian, M. Antimicrobial activity of resveratrol analogues. Molecules 2014, 19, 7679–7688.
- Suga, T.; Ohta, S.; Munesada, K.; Ide, N.; Kurokawa, M.; Shimizu, M.; Ohta, E. Endogenous pine wood nematicidal substances in pines, Pinus massoniana, strobus and P. palustris. Phytochemistry 1993, 33, 1395–1401.
- Torres, P.; Avila, J.G.; de Vivar, A.R.; Garcı́a, A.M.; Marı́n, J.C.; Aranda, E.; Céspedes, C.L. Antioxidant and insect growth regulatory activities of stilbenes and extracts from Yucca periculosa. Phytochemistry 2003, 64, 463–473.
- Liu, Y.Q.; Li, X.J.; Zhao, C.Y.; Lu, Y.; Li, W.Q.; Liu, Z.L.; Feng, G.; Yang, L. Synthesis and insect antifeedant activity of stilbene derivatives against Brontispa longissima Med. Chem. Res. 2013, 22, 2196–2206.
- Hansen, S.C.; Stolter, C.; Imholt, C.; Jacob, J. Plant secondary metabolites as rodent repellents: A systematic review. Chem. Ecol. 2016, 42, 970–983.
- Bryant, J.P.; Wieland, G.D.; Reichardt, P.B.; Lewis, V.E.; McCarthy, M.C. Pinosylvin methyl ether deters snowshoe hare feeding on green alder. Science 1983, 222, 1023–1025.
- Clausen, T.P.; Reichardt, P.B.; Bryant, J.P. Pinosylvin and pinosylvin methyl ether as feeding deterrents in green alder. Chem. Ecol. 1986, 12, 2117–2131.
- Virjamo, V.; Julkunen-Tiitto, R.; Henttonen, H.; Hiltunen, E.; Karjalainen, R.; Korhonen, J.; Huitu, O. Differences in vole preference, secondary chemistry and nutrient levels between naturally regenerated and planted Norway spruce seedlings. Chem. Ecol. 2013, 39, 1322–1334.
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant 2002, 7, 405–410.
- Hasan, M.; Bae, H. An overview of stress-induced resveratrol synthesis in grapes: Perspectives for resveratrol-enriched grape products. Molecules 2017, 22, 294.
- Gülçin, İ. Antioxidant properties of resveratrol: A structure–activity insight. Food Sci. Emerg. Technol. 2010, 11, 210–218.
- Banez, M.J.; Geluz, M.I.; Chandra, A.; Hamdan, T.; Biswas, O.S.; Bryan, N.S.; Von Schwarz, E.R. A systemic review on the antioxidant and anti-inflammatory effects of resveratrol, curcumin, and dietary nitric oxide supplementation on human cardiovascular health. Res. 2020, 78, 11–26.
- Vervandier-Fasseur, D.; Latruffe, N. The potential use of resveratrol for cancer prevention. Molecules 2019, 24, 4506.
- Ahmadi, R.; Ebrahimzadeh, M.A. Resveratrol–A comprehensive review of recent advances in anticancer drug design and development. J. Med. Chem. 2020, 200, 112356.
- Yang, M.F.; Yao, X.; Chen, L.M.; Gu, J.Y.; Yang, Z.H.; Chen, H.F.; Zheng, X.; Zheng, Z.T. Synthesis and biological evaluation of resveratrol derivatives with anti-breast cancer activity. Pharm. (Weinheim) 2020, 353, e2000044.
- Sun, A.Y.; Wang, Q.; Simonyi, A.; Sun, G.Y. Resveratrol as a therapeutic agent for neurodegenerative diseases. Neurobiol. 2010, 41, 375–383.
- Cheng, C.K.; Luo, J.Y.; Lau, C.W.; Chen, Z.Y.; Tian, X.Y.; Huang, Y. Pharmacological basis and new insights of resveratrol action in the cardiovascular system. J. Pharmacol. 2020, 177, 1258–1277.
- Seo, Y.; Park, J.; Choi, W.; Ju Son, D.; Sung Kim, Y.; Kim, M.K.; Yoon, B.E.; Pyee, J.; Hong, J.T.; Go, M.Y.; et al. Antiatherogenic effect of resveratrol attributed to decreased expression of ICAM-1 (Intercellular adhesion Molecule-1) mechanistic link from focal adhesion to monocyte adhesion. Thromb. Vasc. Biol. 2019, 39, 675–684.
- Li, J.; Zhang, C.X.; Liu, Y.M.; Chen, K.L.; Chen, G. A comparative study of anti-aging properties and mechanism: Resveratrol and caloric restriction. Oncotarget 2017, 8, 65717.
- Zhu, X.; Wu, C.; Qiu, S.; Yuan, X.; Li, L. Effects of resveratrol on glucose control and insulin sensitivity in subjects with type 2 diabetes: Systematic review and meta-analysis. Metab. 2017, 14, 60.
- Zu, Y.; Wang, S. Resveratrol-loaded liposomes: Browning subcutaneous white adipose tissue for combating obesity in C57BL/6 J mice. Dev. Nutr. 2020, 4, 1709–1709.
- Kim, H.; Seo, K.H.; Yokoyama, W. Chemistry of pterostilbene and its metabolic effects. Agr. Food Chem. 2020, doi:10.1021/acs.jafc.0c00070.
- Xu, H.; Deng, R.; Li, E.T.; Shen, J.; Wang, M. Pinosylvin provides neuroprotection against cerebral ischemia and reperfusion injury through enhancing PINK1/Parkin mediated mitophagy and Nrf2 pathway. Funct. Foods 2020, 71, 104019.
- Piotrowska, H.; Kucinska, M.; Murias, M. Biological activity of piceatannol: Leaving the shadow of resveratrol. Res. 2012, 750, 60–82.
- Yu, B.; Jiang, Y.; Zhang, B.; Yang, H.; Ma, T. Resveratrol dimer trans-ε-viniferin prevents rotaviral diarrhea in mice by inhibition of the intestinal calcium-activated chloride channel. Res. 2018, 129, 453–461.
- Nivelle, L.; Aires, V.; Rioult, D.; Martiny, L.; Tarpin, M.; Delmas, D. Molecular analysis of differential antiproliferative activity of resveratrol, epsilon viniferin and labruscol on melanoma cells and normal dermal cells. Food Toxicol. 2018, 116, 323–334.
- Global Resveratrol Market Research Report 2020. Industry Research. 2020. Available online: https://www.industryresearch.co/global-resveratrol-market-15064120 (accessed on 01-Dec-2020).
- Huang, H.; Liu, R.; Ou, W. A mini review on the chemical synthesis of resveratrol. Mini Rev. Org. Chem. 2020, 17, 546–558.
- Lv, M.; Zhang, Y.; Wang, F.; Zhang, S.; Xu, H. Non-food renewable and bioactive forest products for pest management: Valuation of agricultural properties of podophyllotoxin analogs derived from Podophyllum hexandrum as botanical pesticides. Crops Prod. 2020, 153, 112608.
- Donati, L.; Ferretti, L.; Frallicciardi, J.; Rosciani, R.; Valletta, A.; Pasqua, G. Stilbene biosynthesis and gene expression in response to methyl jasmonate and continuous light treatment in Vitis vinifera Malvasia del Lazio and Vitis rupestris Du Lot cell cultures. Physiol. Plant. 2019, 166, 646–662.
- Huber, R.; Marcourt, L.; Schnee, S.; Michellod, E.; Wolfender, J.L.; Gindro, K.; Queiroz, E.F. Biotransformations with the enzymatic secretome of Botrytis cinerea combined with organic solvents for the generation of novel complex stilbene derivatives. Planta Med. 2019, 85, 1446–1447.
- Wang, C.; Zhi, S.; Liu, C.; Xu, F.; Zhao, A.; Wang, X.; Ren, Y.; Li, Z.; Yu, M. Characterization of stilbene synthase genes in mulberry (Morus atropurpurea) and metabolic engineering for the production of resveratrol in Escherichia coli. Agric. Food Chem. 2017, 65, 1659–1668.
- He, Q.; Szczepańska, P.; Yuzbashev, T.; Lazar, Z.; Ledesma-Amaro, R. De novo production of resveratrol from glycerol by engineering different metabolic pathways in Yarrowia lipolytica. Eng. Commun. 2020, 11, e00146.
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452.
- Emiliani, G.; Fondi, M.; Fani, R.; Gribaldo, S. A horizontal gene transfer at the origin of phenylpropanoid metabolism: A key adaptation of plants to land. Direct 2009, 4, 1–12.
- Lv, C.; Zhao, G.; Ning, Y. Interactions between plant proteins/enzymes and other food components, and their effects on food quality. Rev. Food Sci. Nutr. 2017, 57, 1718–1728.
- Havir, E.A.; Reid, P.D.; Marsh, H.V. L-phenylalanine ammonia-lyase (maize) evidence for a common catalytic site for L-phenylalanine and L-tyrosine. Plant Physiol. 1971, 48, 130–136.
- Rösler, J.; Krekel, F.; Amrhein, N.; Schmid, J. Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol. 1997, 113, 175–179.
- Barros, J.; Serrani-Yarce, J.C.; Chen, F.; Baxter, D.; Venables, B.J.; Dixon, R.A. Role of bifunctional ammonia-lyase in grass cell wall biosynthesis. Plants 2016, 2, 1–9.
- Barros, J.; Dixon, R.A. Plant phenylalanine/tyrosine ammonia-lyases. Trends Plant Sci. 2020, 25, 66–79.
- Ferrer, J.L.; Austin, M.B.; Stewart, C., Jr.; Noel, J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370.
- Rupprich, N.; Hildebrand, H.; Kindl, H. Substrate specificity in vivo and in vitro in the formation of stilbenes. Biosynthesis of rhaponticin. Biochem. Biophys. 1980, 200, 72–78.
- Schöppner, A.; Kindl, H. Purification and properties of a stilbene synthase from induced cell suspension cultures of peanut. Biol. Chem. 1984, 259, 6806–6811.
- Bais, A.J.; Murphy, P.J.; Dry, I.B. The molecular regulation of stilbene phytoalexin biosynthesis in Vitis vinifera during grape berry development. Plant Biol. 2000, 27, 425–433.
- Samappito, S.; Page, J.E.; Schmidt, J.; De-Eknamkul, W.; Kutchan, T.M. Aromatic and pyrone polyketides synthesized by a stilbene synthase from Rheum tataricum. Phytochemistry 2003, 62, 313–323.
- Fliegmann, J.; Schröder, G.; Schanz, S.; Britsch, L.; Schröder, J. Molecular analysis of chalcone and dihydropinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related enzyme activities in stressed plants. Plant Mol. Biol. 1992, 18, 489–503.
- Kodan, A.; Kuroda, H.; Sakai, F. A stilbene synthase from Japanese red pine (Pinus densiflora): Implications for phytoalexin accumulation and down-regulation of flavonoid biosynthesis. Natl. Acad. Sci. USA 2002, 99, 3335–3339.
- Raiber, S.; Schröder, G.; Schröder, J. Molecular and enzymatic characterization of two stilbene synthases from Eastern white pine (Pinus strobus) A single Arg/His difference determines the activity and the pH dependence of the enzymes. FEBS Lett. 1995, 361, 299–302.
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Prod. Rep. 2003, 20, 79–110.
- Vannozzi, A.; Dry, I.B.; Fasoli, M.; Zenoni, S.; Lucchin, M. Genome-wide analysis of the grapevine stilbene synthase multigenic family: Genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 2012, 12, 130.
- Tropf, S.; Lanz, T.; Rensing, S.A.; Schröder, J.; Schröder, G. Evidence that stilbene synthases have developed from chalcone synthases several times in the course of evolution. Mol. Evol. 1994, 38, 610–618.
- Parage, C.; Tavares, R.; Rety, S.; Baltenweck-Guyot, R.; Poutaraud, A.; Renault, L.; Heintz, D.; Lugan, R.; Marais, G.A.; Aubourg, S.; et al. Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol. 2012, 160, 1407–1419.
- Flores-Sanchez, I.J.; Verpoorte, R. Plant polyketide synthases: A fascinating group of enzymes. Plant Physiol. Biochem. 2009, 47, 167–174.
- Pandith, S.A.; Ramazan, S.; Khan, M.I.; Reshi, Z.A.; Shah, M.A. Chalcone synthases (CHSs): The symbolic type III polyketide synthases. Planta 2020, 251, 15.
- Austin, M.B.; Bowman, M.E.; Ferrer, J.L.; Schröder, J.; Noel, J.P. An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Biol. 2004, 11, 1179–1194.
- Li, H.; Liang, J.; Chen, H.; Ding, G.; Ma, B.; He, N. Evolutionary and functional analysis of mulberry type III polyketide synthases. BMC Genom. 2016, 17, 540.
- Schröder, G.; Brown, J.W.; Schröder, J. Molecular analysis of resveratrol synthase: cDNA, genomic clones and relationship with chalcone synthase. J. Biochem. 1988, 172, 161–169.
- Melchior, F.; Kindl, H. Coordinate-and elicitor-dependent expression of stilbene synthase and phenylalanine ammonia-lyase genes in Vitis Optima. Arch. Biochem. Biophys. 1991, 288, 552–557.
- Preisig-Müller, R.; Schwekendiek, A.; Brehm, I.; Reif, H.J.; Kindl, H. Characterization of a pine multigene family containing elicitor-responsive stilbene synthase genes. Plant Mol. Biol. 1999, 39, 221–229.
- Warren, R.L.; Keeling, C.I.; Yuen, M.M.; Raymond, A.; Taylor, G.A.; Vandervalk, B.P.; Mohamadi, H.; Paulino, D.; Chiu, R.; Jackman, S.D.; et al. Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J. 2015, 83, 189–212.
- Hammerbacher, A.; Ralph, S.G.; Bohlmann, J.; Fenning, T.M.; Gershenzon, J.; Schmidt, A. Biosynthesis of the major tetrahydroxystilbenes in spruce, astringin and isorhapontin, proceeds via resveratrol and is enhanced by fungal infection. Plant Physiol. 2011, 157, 876–890.
- Christine, K.Y.; Springob, K.; Schmidt, J.; Nicholson, R.L.; Chu, I.K.; Yip, W.K.; Lo, C. A stilbene synthase gene (SbSTS1) is involved in host and nonhost defense responses in Sorghum. Plant Physiol. 2005, 138, 393–401.
- Zhu, F.; Han, J.; Liu, S.; Chen, X.; Varshney, R.K.; Liang, X. Cloning, expression pattern analysis and subcellular localization of resveratrol synthase gene in peanut (Arachis hypogaea). Am. J. Plant Sci. 2014, 5, 3619–3631.
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al.The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556.
- Sharma, I.; Kumari, N.; Sharma, V. Defense gene expression in Sorghum bicolor against Macrophomina phaseolina in leaves and roots of susceptible and resistant cultivars. Plant Interact. 2014, 9, 315–323.
- Lee, Y.G.; Choi, S.C.; Kang, Y.; Kim, K.M.; Kang, C.S.; Kim, C. Constructing a reference genome in a single lab: The possibility to use oxford nanopore technology. Plants 2019, 8, 270.
- Sparvoli, F.; Martin, C.; Scienza, A.; Gavazzi, G.; Tonelli, C. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera). Plant Mol. Biol. 1994, 24, 743–755.
- Dai, R.; Ge, H.; Howard, S.; Qiu, W. Transcriptional expression of stilbene synthase genes are regulated developmentally and differentially in response to powdery mildew in Norton and Cabernet Sauvignon grapevine. Plant Sci. 2012, 197, 70–76.
- Shi, J.; He, M.; Cao, J.; Wang, H.; Ding, J.; Jiao, Y.T.; Li, R.M.; He, J.; Wang, D.; Wang, Y. The comparative analysis of the potential relationship between resveratrol and stilbene synthase gene family in the development stages of grapes (Vitis quinquangularis and Vitis vinifera). Plant Physiol. Biochem. 2014, 74, 24–32.