Dynamic interactions between gut microbiota and a host’s innate and adaptive immune systems play key roles in maintaining intestinal homeostasis and inhibiting inflammation. The gut microbiota metabolizes proteins and complex carbohydrates, synthesize vitamins, and produce an enormous number of metabolic products that can mediate cross-talk between gut epithelial and immune cells. As a defense mechanism, gut epithelial cells produce a mucosal barrier to segregate microbiota from host immune cells and reduce intestinal permeability. An impaired interaction between gut microbiota and the mucosal immune system can lead to an increased abundance of potentially pathogenic gram-negative bacteria and their associated metabolic changes, disrupting the epithelial barrier and increasing susceptibility to infections. Gut dysbiosis, or negative alterations in gut microbial composition, can also dysregulate immune responses, causing inflammation, oxidative stress, and insulin resistance. Over time, chronic dysbiosis and the translocation of bacteria and their metabolic products across the mucosal barrier may increase prevalence of type 2 diabetes, cardiovascular disease, inflammatory bowel disease, autoimmune disease, and a variety of cancers.
- gut microbiota,immune system,gut microbiota meta
1. Gut microbial dysbiosis and the host immune dysregulation.
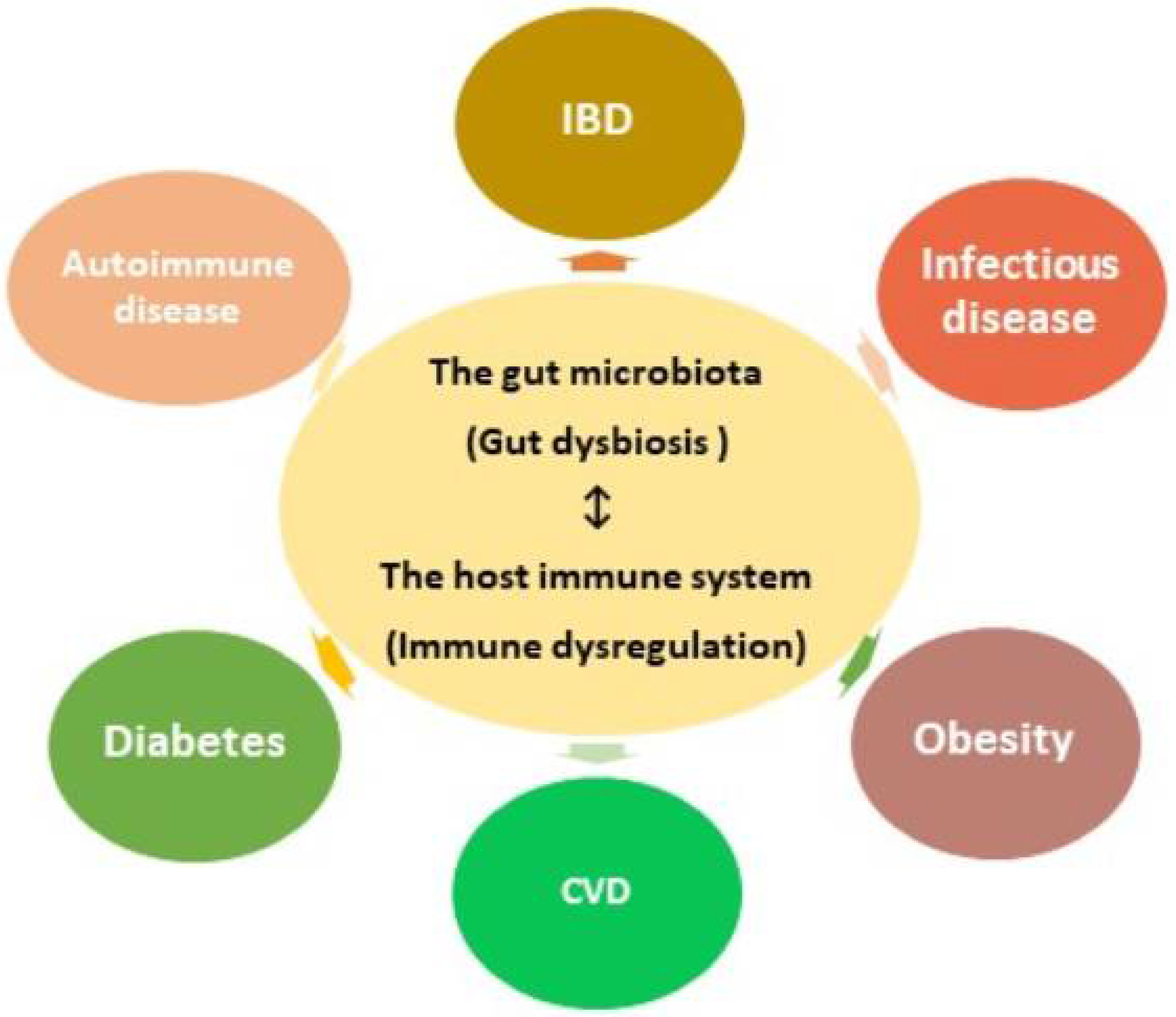
The mucosal barrier secreted by gut epithelial cells acts as a defense mechanism, segregating microbes from host immune cells and reducing intestinal permeability. Disrupting the epithelial barrier increases susceptibility to infection and the displacement of microbial metabolites into the host. Gut dysbiosis, or negative alterations in the gut microbial composition, not only reduces the integrity of the mucosal barrier, but also dysregulates immune responses, causing inflammation, oxidative stress, and insulin resistance. Over time, chronic gut dysbiosis and the passage of bacteria and their metabolic products through the mucosal barrier can increase the prevalence of a variety of diseases.
3.1. Gut Microbiota and Chronic Low-grade Inflammation on Type 2 Diabetes (T2DM) and CVD
1.1. Gut Microbiota and Chronic Low-grade Inflammation on Type 2 Diabetes (T2DM) and CVD
A number of studies provide evidence supporting associations between gut dysbiosis, T2DM (non-insulin-dependent), and CVD [80–83][1][2][3][4]. One cause of T2DM is reduced insulin function in the liver, skeletal muscle, and adipose tissue [84][5]. Insulin is a hormone that acts as a key mediator regulating glucose homeostasis and lipid metabolism. Insulin has varied functions, including regulating gene expression, stimulating metabolite uptake into cells, altering enzymatic activity, and maintaining energy homeostasis. Insulin also enhanced glucose uptake by promoting relocation of the principal glucose transporter (GLUT4) to the cell membrane in skeletal muscle and other tissues [85][6]. In skeletal muscle, GLUT4 functions in insulin-dependent glucose disposal [86,87][7][8]. As a result, poor insulin signaling leads to impaired glucose disposal in skeletal muscle, markedly seen in T2DM patients. Although the precise cause of resistance to insulin in skeletal muscle remains elusive, increased intramyocellular lipid content and free fatty acid (FFA) metabolites play an essential role [88,89][9][10]. Increased circulating FFAs decreased the sensitivity of skeletal muscle to insulin via increased levels of fatty acyl-CoA, ceramide, and other intracellular lipid products [90,91][11][12]. These lipid metabolites activated the serine/threonine kinase protein kinase C-θ (PKC-θ), inhibiting downstream insulin signaling. The inhibition of insulin signaling in the liver induced the activity of important gluconeogenic enzymes, resulting in insulin resistance and increased hepatic glucose production [92][13]. Reduced activity in hormone sensitive lipase and the anti-lipolytic effect inhibited FFA efflux from adipocytes by adipose tissue insulin signaling. Disruption of these processes occurred in both insulin resistance and deficiency, and elevating fasting and postprandial glucose and lipid levels, major causes of T2DM and CVD [93][14].
A well-known gut microbiota related pathway is trimethylamine–N-oxide (TMAO) metabolism. Dietary phospholipids (lecithin, choline, and carnitine) are anaerobically metabolized by bacteria, yielding the products ethanolamine and trimethyl amine (TMA). Gut microbial TMA lyases cleave the C-N bond of dietary phospholipids, producing TMA as a waste product. These nutrients are all substrates of gut bacteria and are metabolized to form the products TMA/TMAO. Conversion of TMA to TMAO proceeds through an oxidation pathway moderated by host hepatic enzymes, including flavin monooxygenases (FMOs), to which TMA is exposed via portal circulation, forming TMAO. Elevated TMAO has been correlated with incidence of CVD and increased danger of myocardial infarction (MI), stroke, revascularization, and oxidation, and has also been used to predict major negative cardiac outcomes over a three-year period [94][15]. While increased circulating levels of betaine, choline, and carnitine have been correlated with future risk of negative cardiac outcome, their prognostic value was limited without a similar elevation in TMAO levels. Wang et al. (2011) [95][16] demonstrated an increased risk of atherosclerosis associated with plasma choline, TMAO, and betaine levels in mice and humans. Further studies show TMAO levels are associated with abundances of the genera Prevotella and Bacteroides, and applied dietary TMAO supplementation promoted decreased total cholesterol absorption in mouse models [96][17].
Numerous studies support the association between chronic low-grade inflammation, T2DM, and CVD [97][18]. Insulin resistance often results in compensatory hyperinsulinemia, one cause of metabolic disruptions commonly seen in metabolic syndrome, a precursor to CVD [87][8]. Moreover, low-grade inflammation is related to the presence of excessive visceral fat, and can lead to insulin resistance, greater macrophage infiltration, and the subsequent manufacture of pro-inflammatory adipokines. This chronic low-grade inflammation via the suppression of the insulin-signaling pathway in peripheral tissues was associated with T2DM and the development of CVD [98][19]. The excess of inflammatory factors such as TNF-α, IL-6, and IL-1 are associated with reduced insulin action, and help modulate interactions linking the immune system and insulin signaling.
The gut dysbiosis associated with T2DM has been well-documented, including decreased populations of butyrate-producing bacteria and increased resistance to oxidative stresses characteristic of opportunistic pathogens [99,100][20][21]. SCFAs increased anti-inflammatory responses in the host body, however, T2DM patients have shown significantly fewer SCFA-producing bacteria and significantly increased LPS levels [97,101][18][22]. LPS leakage into the body from gram-negative microbes is a contributing factor for low-grade inflammation, inducing the release of proinflammatory molecules. Systemic inflammation was strongly correlated with high levels of triglycerides and decreased serum HDL, increased blood pressure, and increased fasting glucose [101[22][23],102], molecules that increased intestinal epithelial inflammation and translocation through the gut barrier. LPS receptors were found to be critical mediators that potentially activate underlying insulin resistance. In pancreatic islets, TLR4 increased proinflammatory cytokine levels, limiting macrophage and beta-cell viability and reducing their functionality [103][24]. Moreover, the activation of TLR4 was directly related to lower mRNA levels of pancreas-duodenum homeobox-1 (PDX-1), lower insulin mRNA and protein levels, and reduced insulin-induced glucose secretion. Also, LPS upregulated nuclear factor-κB (NF-κB) levels and initiated mitogen-activated protein kinase (MAPK)-mediated signaling in lipocytes. The anti-inflammatory effects of SCFAs likely arise from a balance, reducing the levels of proinflammatory mediators and promoting anti-inflammatory cytokine production. Butyrate and propionate not only reduced the levels of proinflammatory cytokines IL-6 and TNF-α in human adipose tissue, but also stimulated the secretion of the anti-inflammatory cytokine IL-10 by monocytes interactions with bacteria. SCFAs decreased low-grade inflammation through their ability to modulate both leukocytes and adipocytes, decreasing inflammatory cytokine and chemokine levels [104][25]. Other studies have demonstrated that SCFAs ameliorated the LPS-induced neutrophil release of TNF-α. Accordingly, SCFAs significantly reduce expression of several inflammation associated cytokines and chemokines in adipocytes, decreasing the risk of T2DM and CVD.
Another important function of SCFAs in T2DM is binding GPCRs, triggering various downstream effects. SCFAs stimulate healthy glucose tolerance through the release of gut-derived incretin hormones such as Glucagon-like peptide-1 (GLP-1). This hormone is primarily synthesized by intestinal L-cells and released into circulation during meal ingestion. GLP-1 stimulates weight loss by reducing glucagon secretion and hepatic gluconeogenesis, while increasing insulin response, improving satiety. The utility of GLP-1 for treatment purposes has been noted, as several therapeutic agents enhancing GLP-1 are available and successfully used to monitor blood glucose levels in patients with T2D, as well as providing improved body weight management in obese patients. Intravenous GLP-1 is efficient in promoting insulin release and decreasing hyperglycemia in T2DM patients. Furthermore, GLP-1 mimetics and a dipeptidyl peptidase-4 inhibitor are commonly used therapeutic approaches in clinics [105][26]. In particular, injectable GLP-1 mimetics are associated with decreased concentrations of fasting and postprandial glucose, decreased hemoglobin A1c (HbA1c), and significant weight loss.
Among SCFAs, propionate and acetate strongly stimulated GLP-1 release, while butyrate provided a weaker stimulation [106][27]. However, improved GLP-1 synthesis and secretion has been noted in the human L-cell line treated with butyrate. Further, the improvements noted after administration of the prebiotic showed the association with increased secretion of GLP-1, reduced insulin resistance, and decreased body weight gain [107][28]. Mechanisms by which GLP-1 secretion is improved after SCFA treatment are still a matter of debate. The limited response of GLP-1 to SCFA treatment in the GLUTag cell line coincides with low levels of GPR43 [106][27]. Extended exposure to a high-fat diet generated lower levels of insulin in GPR43 knockout mice, as opposed to wild-type controls [108]. Conversely, GPR41 may mediate the molecular interactions between butyrate and human L-cells, as increases in butyrate induced GLP-1 expression were correlated with high levels of GPR41. Additionally, the increased GLP-1 secretion stimulated by butyrate was weakened in GPR41 knockout mice [108][29]. SCFAs are associated with regulation of insulin levels via GLP-1 expression, and result in improvement of metabolic functions in T2DM. The regulation of blood glucose concentrations exerted by SCFAs occur through multiple mechanisms, including reduced insulin resistance from decreased inflammation, the secretion of GLP-1 and subsequent insulin release, and increased beta-cell activity promoting glucose homeostasis.
3.2. Gut Microbiota and Inflammatory Bowel Disease (IBD)
1.2. Gut Microbiota and Inflammatory Bowel Disease (IBD)
Ulcerative colitis (UC) and Crohn’s disease (CD) are chronic conditions of the GI tract characterized by increased levels of inflammation resulting from impaired interactions between microbiota in the gut and the intestinal immune cells. These complex inflammatory bowel diseases (IBDs) result from a combination of negative environmental stimuli, gut dysbiosis, and host genetics. Major shifts in gut microbial composition, including increases in facultative anaerobic pathogens and decreases in obligate anaerobic producers of SCFAs commonly occur in IBD patients [109][30]. IBD is characterized by a breakdown in interactions between the resident microbial population and host immune responses. While no single pathogenic species has been etiologically linked to IBD, changes to microbial abundances in the dysbiotic gut play critical roles in the persistent inflammation found observed [97,110,111][31][32].
The association of gut microbiota in IBD onset and progression has been demonstrated in both murine models and IBD patients. Initial studies performed by Rutgeerts (1991) [112] [33] and D’Haens (1998) [113][34] demonstrated the influence of gut microbes in IBD using clinical experiments that showed fecal stream diversion ameliorated the symptoms of CD, and that inflammation resulted when luminal microbiota were subsequently transferred to the terminal ileum. In CD patients, increased gut permeability promotes bacterial translocation through the intestinal mucosa. In healthy individuals, the gut barrier consists of a layer of tightly connected epithelial cells surrounded by a system of tight junction strands from the claudin protein family. In mild to moderate CD, gut barrier dysfunctions resulted mostly from the increased levels of claudin 2, while the expression and redistribution of claudins 5 and 8 was decreased, leading to discontinuous tight junctions [114,115][35][36]. In IBD patients, gut dysbiosis occurs due to a combination of colonization by enteric pathogens and host inflammatory responses. Pathogens can use inflammation to translocate across the intestinal mucosal barrier, subverting host inflammatory responses. Salmonella typhimurium, for example, induced inflammation responses that altered the composition of the gut microbiota and promoted its own growth in mouse models [116,117][37][38]. Another pathogen, Campylobacter jejuni, is a common bacterial pathogen in humans. In dextran sulfate sodium (DSS)-induced colitis mouse models, non-inflammatory infection with Campylobacter jejuni decreased the total number of colonic bacteria, but DSS-induced inflammation encouraged overgrowth of Enterobacteriaceae, with both conditions required for maximal gut dysbiosis [118,119][39][40]. Harnessing microbiota to influence host immunity is a meaningful treatment strategy for patients with IBD.
Notably, numerous studies have identified changes in bacteria associated with CD, particularly reduced abundances of the phylum Firmicutes and increases in Proteobacteria [114,120]. Specifically, Clostridium clusters IV and XIV have been observed in decreased amounts in IBD patients in contrast to non-IBD controls, revealing that their absence may diminish the gut of anti-inflammatory immunomodulatory ligands and metabolites. Both UC and CD gut microbiota show reduced taxonomic diversity in comparison to healthy controls, along with increased abundance in the phyla Proteobacteria, and decreased abundance of the phyla Firmicutes. The quantity of bacteria from the Clostridia family are also commonly altered, with decreases in Roseburia and Faecalibacterium and increases in Ruminococcus and the Enterobacteriaceae family observed in IBD patients [120,121][41][42]. Together, these findings suggest that IBD is ultimately linked to inflammation that may be largely associated with microbially derived or modified metabolites.
Mutations of the NOD2 gene increase susceptibility to CD. PRRs, including TLRs, NOD-like receptors (NLRs), and others, recognize pathogen-associated molecular patterns found in microbial pathogens, a critical step in the innate immune response. IBD genetic association studies have discovered functionally relevant polymorphisms in several innate immune genes. Of those identified, mutations in the receptor NOD2 conferred the most risk. NOD2 is an intracellular sensor, detecting microbial pathogens and helping maintain epithelial defense, detecting peptidoglycans through the recognition of muramyl dipeptide [121][42]. NOD2 activates nuclear factor NF-kB, an inhibitory protein and intracellular receptor that binds portions of microbial pathogens. After binding with intracellular muramyl dipeptide (MDP), NOD2 recruits the adaptor protein RIP2, activating NF-κB and initiating a pro-inflammatory response. One of the first genes associated with susceptibility to CD was NOD2, and NOD2 loss of function mutations frequently occur in IBD patients. In one study, NOD2-/- mice exhibited a large decrease in the number of intestinal intraepithelial lymphocytes (IELs), and the remaining IELs showed increased apoptosis and little proliferation. NOD2 signaling recognized gut bacteria and stimulated IL-15 production, maintaining IEL abundances and providing a clue linking NOD2 variation to increased incidence of CD and reduced innate immunity [121][42].
3.3. Gut Microbiota and Systemic Lupus Erythematosus (SLE)
1.3. Gut Microbiota and Systemic Lupus Erythematosus (SLE)
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease that exhibits increased levels of autoantibodies and reduced tolerance to self-antigens, mirroring dysregulation in both the innate and adaptive immune systems. While the pathogenesis is not fully understood, a combination of gut microbiota, host genetics, and environmental factors contribute to SLE [122][43]. The associated loss of immune tolerance can be stimulated by altered gut microbial composition, with both experimental and clinical models providing evidence of the strong relationship between gut dysbiosis and SLE [123–125][44][45][46].
Gut microbiota can stimulate an immune response targeting the host for any of several reasons. In normal cell apoptotic conditions, the host immune system does not involve the release of nuclear antigens. In SLE, however, external stimuli such as bacterial infection, toxins, or ultraviolet (UV) light induce DNA damage and keratinocyte apoptosis. The resulting prolonged autoantigen exposure increased stimulation of host immune cell responses, inducing T-cell activation via suppression of Treg cells in a type I interferon (IFN)-dependent manner [126][47]. The type I IFN pathway is stimulated by nucleic acid containing cell fragments through recognition by nucleic acid recognition receptors TLR7 or TLR9 [127,128][48][49]. Type I IFNs and other cytokines encourage the development and survival of B-cells, inducing the B-cell hyperactivity characteristic of SLE. B-cells create autoantibodies that target self-antigens with high-affinity, causing inflammation and tissue damage. Autoantibodies and immune complexes (ICs) mitigate the inflammatory damage by stimulating complement cascades and interacting with Fc receptors on inflammatory cells. Fc receptors are a heterogeneous collection of hematopoietic cell surface glycoproteins used by immune system effector cells to increase antibody–antigen interactions. These receptors are involved in a host of immune outcomes including phagocytosis, cytotoxicity, inflammatory chemokine and cytokine levels, B-cell activity, and IC clearance. The expression and function of the Fc region in IgG (FcγR) are altered in SLE. FcγR stimulates the manufacture of autoantibodies, resulting in inflammation and IC handling, contributing to the onset and progression of SLE. Altered or delayed removal of ICs containing autoantibodies lead to their accumulation in various tissues, causing inflammation and cellular damage by engaging FcγRs and complement cascades [129,130][50][51].
An increased abundance of follicular Th cells and deficiencies in Treg cells have also been associated with SLE pathogenesis. Scalapino et al. (2006) [131][52] demonstrated with adoptive transfer experiments that Treg cells hindered disease advancement and increased the life span of lupus-prone mice. CD4+ Treg cells preserve auto-tolerance by inhibiting autoreactive immune cells, leading to the hypothesis that Treg cell defects contribute to the onset and severity of SLE. Proinflammatory cytokines were secreted by activated T-cells, which induced B-cell secretion of autoantibodies, pushing both innate and adaptive immune responses closer to autoimmunity [36,132][53]. In germ-free murine models, T-cell compositions in the gut were affected, displaying reduced responses of Th17 and Treg in the small intestine and colonic lamina propria, respectively [133,134][54][55]. Gut microbiota has been associated with the altered composition of Th17 and Treg cells in SLE patients [125][46]. In patients with SLE, the gut microbial composition was significantly enriched in several genera, including Klebsiella, Rhodococcus, Eggerthella, Eubacterium, Prevotella, and Flavonifractor. In contrast, the abundances of Dialister and Pseudobutyrivibrio and the Firmicutes/Bacteroidetes ratio have been suppressed in SLE patients [123][44]. It remains unclear whether the observed changes in commensal bacteria are a consequence of the disease process or if gut dysbiosis contributes to SLE onset. However, manipulation of gut microbiota in murine models with the antibiotic treatment provided further evidence of gut microbiota influences on systemic immune homeostasis [135,136][56][57].
Furthermore, studies showed significant reductions of Lactobacillaceae and increased abundances of Lachnospiraceae in female lupus-prone mice [137][58]. Lactobacillus, a genus in the Lactobacillaceae family, is a common resident microbiota in human GI tracts. Some Lactobacillus species are marketed as probiotics due to their anti-inflammatory properties, and the decreased abundance of Lactobacillus spp. was most pronounced prior to disease onset. Lactobacillus treatment decreases IL-6 and increases IL-10 production, creating an anti-inflammatory environment in the epithelium. In circulation, Lactobacillus supplementation promoted IL-10 and reduced levels of IgG2a, a significant immune repository in MRL/lpr mice. Thus, Lactobacillus may have a preventive influence in lupus pathogenesis [138][59]. However, Lactobacillus played a divergent part in alternative lupus murine studies, where the abundance of Lactobacillus spp. correlated strongly with systemic autoimmunity and negatively with renal function in NZB/W F1 mice [124, 139][45][60]. Consideration of the gut microbiota in murine models and human clinical cases of lupus have offered novel insights on the role of bacteria in SLE onset and progression.
Opposing correlations were reported between pro-and anti-inflammatory free fatty acids (FFAs) and endothelial markers in lupus patients, supporting the connection linking gut microbiota and host metabolism in the etiology of SLE [140]. The altered production of SCFAs stemming from intestinal dysbiosis highlights the role of the gut in maintaining serum FFA levels, further pointing to SCFAs as one potential mechanism for cross-talk linking the host metabolism and gut microbiota [140][61]. Additionally, five perturbed metabolic pathways were identified in feces of SLE patients, including the metabolism of tryptophan, nitrogen, thiamine, and cyanoamino acids, as well as aminoacyl-tRNA biosynthesis [141][62]. The aforementioned metabolites can potentially be used as biomarkers for SLE, as long as the effects of potential cofounders, such as smoking, diet, medication, and co-morbidities are considered [142][63]. Given the role of metabolites in autoimmune diseases, one proposed therapeutic strategy is to promote the differentiation of CD4+ T-cells into Treg cells while avoiding differentiation into Th1 and Th17 cells by inducing the fermentation of SCFAs by gut bacteria. The active proliferation of SCFA-producing gut microbes have been stimulated using appropriate types of dietary fiber [143][64], prebiotics such as fructo-oligosaccharides [144,145][65][66], or probiotics [146][67], though direct oral supplementation with SCFAs may be problematic as they are metabolized early in the intestinal tract [147][68].
References
- Everard, A.; Cani, P.D. Diabetes, obesity and gut microbiota. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 73–83, doi:10.1016/j.bpg.2013.03.007.
- Miele, L.; Giorgio, V.; Alberelli, M.A.; De Candia, E.; Gasbarrini, A.; Grieco, A. Impact of Gut Microbiota on Obesity, Diabetes, and Cardiovascular Disease Risk. Curr. Cardiol. Rep. 2015, 17, 120, doi:10.1007/s11886-015-0671-z.
- Sanz, Y.; Santacruz, A.; Gauffin, P. Gut microbiota in obesity and metabolic disorders. Proc. Nutr. Soc. 2010, 69, 434–441, doi:10.1017/S0029665110001813.
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845, doi:10.1038/s41467-017-00900-1.
- Yang, Q.; Graham, T.E.; Mody, N.; Preitner, F.; Peroni, O.D.; Zabolotny, J.M.; Kotani, K.; Quadro, L.; Kahn, B.B. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 2005, 436, 356–362, doi:10.1038/nature03711.
- Huang, S.; Czech, M.P. The GLUT4 glucose transporter. Cell Metab. 2007, 5, 237–252, doi:10.1016/j.cmet.2007.03.006.
- Garvey, W.T.; Maianu, L.; Zhu, J.-H.; Brechtel-Hook, G.; Wallace, P.; Baron, A.D. Evidence for defects in the trafficking and translocation of GLUT4 glucose transporters in skeletal muscle as a cause of human insulin resistance. J. Clin. Investig. 1998, 101, 2377–2386.
- de Luca, C.; Olefsky, J.M. Inflammation and insulin resistance. FEBS Lett. 2008, 582, 97–105, doi:10.1016/j.febslet.2007.11.057.
- Krssak, M.; Roden, M. The role of lipid accumulation in liver and muscle for insulin resistance and type 2 diabetes mellitus in humans. Rev. Endocr. Metab. Disord. 2004, 5, 127–134, doi:10.1023/B:REMD.0000021434.98627.dc.
- Snel, M.; Jonker, J.T.; Schoones, J.; Lamb, H.; de Roos, A.; Pijl, H.; Smit, J.W.; Meinders, A.E.; Jazet, I.M. Ectopic fat and insulin resistance: Pathophysiology and effect of diet and lifestyle interventions. Int. J. Endocrinol. 2012, 2012, 983814, doi:10.1155/2012/983814.
- Kurokawa, J.; Arai, S.; Nakashima, K.; Nagano, H.; Nishijima, A.; Miyata, K.; Ose, R.; Mori, M.; Kubota, N.; Kadowaki, T. Macrophage-derived AIM is endocytosed into adipocytes and decreases lipid droplets via inhibition of fatty acid synthase activity. Cell Metab. 2010, 11, 479–492, doi:10.1016/j.cmet.2010.04.013.
- Lampidonis, A.D.; Rogdakis, E.; Voutsinas, G.E.; Stravopodis, D.J. The resurgence of Hormone-Sensitive Lipase (HSL) in mammalian lipolysis. Gene 2011, 477, 1–11, doi:10.1016/j.gene.2011.01.007.
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806, doi:10.1038/414799a.
- Glassing, A.; Dowd, S.E.; Galandiuk, S.; Davis, B.; Chiodini, R.J. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016, 8, 24, doi:10.1186/s13099-016-0103-7.
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584, doi:10.1056/NEJMoa1109400.
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63, doi:10.1038/nature09922.
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585, doi:10.1038/nm.3145.
- Yoo, J.Y.; Kim, S.S. Probiotics and Prebiotics: Present Status and Future Perspectives on Metabolic Disorders. Nutrients 2016, 8, 173, doi:10.3390/nu8030173.
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015, 9, 1–15, doi:10.1038/ismej.2014.99.
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085, doi:10.1371/journal.pone.0009085.
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60, doi:10.1038/nature11450.
- Amar, J.; Burcelin, R.; Ruidavets, J.B.; Cani, P.D.; Fauvel, J.; Alessi, M.C.; Chamontin, B.; Ferriéres, J. Energy intake is associated with endotoxemia in apparently healthy men. Am. J. Clin. Nutr. 2008, 87, 1219–1223, doi:10.1093/ajcn/87.5.1219.
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772, doi:10.2337/db06-1491.
- Lu, Y.C.; Yeh, W.C.; Ohashi, P.S. LPS/TLR4 signal transduction pathway. Cytokine 2008, 42, 145–151, doi:10.1016/j.cyto.2008.01.006.
- Meijer, K.; de Vos, P.; Priebe, M.G. Butyrate and other short-chain fatty acids as modulators of immunity: What relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 715–721, doi:10.1097/MCO.0b013e32833eebe5.
- Drucker, D.J.; Nauck, M.A. The incretin system: Glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet 2006, 368, 1696–1705, doi:10.1016/S0140-6736(06)69705-5.
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes 2012, 61, 364–371, doi:10.2337/db11-1019.
- Yadav, H.; Lee, J.H.; Lloyd, J.; Walter, P.; Rane, S.G. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J. Biol. Chem. 2013, 288, 25088–25097, doi:10.1074/jbc.M113.452516.
- Bjursell, M.; Admyre, T.; Goransson, M.; Marley, A.E.; Smith, D.M.; Oscarsson, J.; Bohlooly, Y.M. Improved glucose control and reduced body fat mass in free fatty acid receptor 2-deficient mice fed a high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2011, 300, E211–E220, doi:10.1152/ajpendo.00229.2010.
- Nagao-Kitamoto, H.; Shreiner, A.B.; Gillilland, M.G., 3rd; Kitamoto, S.; Ishii, C.; Hirayama, A.; Kuffa, P.; El-Zaatari, M.; Grasberger, H.; Seekatz, A.M.; et al. Functional Characterization of Inflammatory Bowel Disease-Associated Gut Dysbiosis in Gnotobiotic Mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 468–481, doi:10.1016/j.jcmgh.2016.02.003.
- Scher, J.U.; Ubeda, C.; Artacho, A.; Attur, M.; Isaac, S.; Reddy, S.M.; Marmon, S.; Neimann, A.; Brusca, S.; Patel, T.; et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015, 67, 128–139, doi:10.1002/art.38892.
- Kaur, N.; Chen, C.C.; Luther, J.; Kao, J.Y. Intestinal dysbiosis in inflammatory bowel disease. Gut Microbes 2011, 2, 211–216, doi:10.4161/gmic.2.4.17863.
- Rutgeerts, P.; Goboes, K.; Peeters, M.; Hiele, M.; Penninckx, F.; Aerts, R.; Kerremans, R.; Vantrappen, G. Effect of faecal stream diversion on recurrence of Crohn's disease in the neoterminal ileum. Lancet 1991, 338, 771–774, doi:10.1016/0140-6736(91)90663-a.
- D'Haens, G.R.; Geboes, K.; Peeters, M.; Baert, F.; Penninckx, F.; Rutgeerts, P. Early lesions of recurrent Crohn's disease caused by infusion of intestinal contents in excluded ileum. Gastroenterology 1998, 114, 262–267, doi:10.1016/s0016-5085(98)70476-7.
- Gassler, N.; Rohr, C.; Schneider, A.; Kartenbeck, J.r.; Bach, A.; Obermüller, N.; Otto, H.F.; Autschbach, F. Inflammatory bowel disease is associated with changes of enterocytic junctions. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 281, G216–G228, doi:10.1152/ajpgi.2001.281.1.G216.
- Miele, L.; Valenza, V.; La Torre, G.; Montalto, M.; Cammarota, G.; Ricci, R.; Masciana, R.; Forgione, A.; Gabrieli, M.L.; Perotti, G.; et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology 2009, 49, 1877–1887, doi:10.1002/hep.22848.
- Winter, S.E.; Thiennimitr, P.; Winter, M.G.; Butler, B.P.; Huseby, D.L.; Crawford, R.W.; Russell, J.M.; Bevins, C.L.; Adams, L.G.; Tsolis, R.M.; et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010, 467, 426–429, doi:10.1038/nature09415.
- Stecher, B.; Robbiani, R.; Walker, A.W.; Westendorf, A.M.; Barthel, M.; Kremer, M.; Chaffron, S.; Macpherson, A.J.; Buer, J.; Parkhill, J.; et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007, 5, 2177–2189, doi:10.1371/journal.pbio.0050244.
- O'Hara, J.R.; Feener, T.D.; Fischer, C.D.; Buret, A.G. Campylobacter jejuni disrupts protective Toll-like receptor 9 signaling in colonic epithelial cells and increases the severity of dextran sulfate sodium-induced colitis in mice. Infect. Immun. 2012, 80, 1563–1571, doi:10.1128/IAI.06066-11.
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 119–129, doi:10.1016/j.chom.2007.06.010.
- Nishino, K.; Nishida, A.; Inoue, R.; Kawada, Y.; Ohno, M.; Sakai, S.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Kawahara, M.; et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 2018, 53, 95–106, doi:10.1007/s00535-017-1384-4.
- Lepage, P.; Häsler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011, 141, 227–236, doi:10.1053/j.gastro.2011.04.011.
- Gladman, D.D.; Ibanez, D.; Urowitz, M.B. Systemic lupus erythematosus disease activity index 2000. J. Rheumatol. 2002, 29, 288–291.
- He, Z.; Shao, T.; Li, H.; Xie, Z.; Wen, C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016, 8, 64, doi:10.1186/s13099-016-0146-9.
- Luo, X.M.; Edwards, M.R.; Mu, Q.; Yu, Y.; Vieson, M.D.; Reilly, C.M.; Ahmed, S.A.; Bankole, A.A. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl. Environ. Microbiol. 2018, 84, doi:10.1128/AEM.02288-17.
- Lopez, P.; de Paz, B.; Rodriguez-Carrio, J.; Hevia, A.; Sanchez, B.; Margolles, A.; Suarez, A. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci. Rep. 2016, 6, 24072, doi:10.1038/srep24072.
- Wolf, S.J.; Estadt, S.N.; Theros, J.; Moore, T.; Ellis, J.; Liu, J.; Reed, T.J.; Jacob, C.O.; Gudjonsson, J.E.; Kahlenberg, J.M. Ultraviolet light induces increased T cell activation in lupus-prone mice via type I IFN-dependent inhibition of T regulatory cells. J. Autoimmun. 2019, 103, 102291, doi:10.1016/j.jaut.2019.06.002.
- Sontheimer, C.; Liggitt, D.; Elkon, K.B. Ultraviolet B Irradiation Causes Stimulator of Interferon Genes-Dependent Production of Protective Type I Interferon in Mouse Skin by Recruited Inflammatory Monocytes. Arthritis Rheumatol. 2017, 69, 826–836, doi:10.1002/art.39987.
- Båve, U.; Magnusson, M.; Eloranta, M.-L.; Perers, A.; Alm, G.V.; Rönnblom, L. FcγRIIa is expressed on natural IFN-α-producing cells (plasmacytoid dendritic cells) and is required for the IFN-α production induced by apoptotic cells combined with lupus IgG. J. Immunol. 2003, 171, 3296–3302, doi:10.4049/jimmunol.171.6.3296.
- Bolland, S.; Ravetch, J.V. Spontaneous autoimmune disease in FcγRIIB-deficient mice results from strain-specific epistasis. Immunity 2000, 13, 277–285, doi:10.1016/S1074-7613(00)00027-3.
- Li, X.; Ptacek, T.S.; Brown, E.E.; Edberg, J.C. Fcγ receptors: Structure, function and role as genetic risk factors in SLE. Genes Immun. 2009, 10, 380–389.
- Scalapino, K.J.; Tang, Q.; Bluestone, J.A.; Bonyhadi, M.L.; Daikh, D.I. Suppression of disease in New Zealand Black/New Zealand White lupus-prone mice by adoptive transfer of ex vivo expanded regulatory T cells. J. Immunol. 2006, 177, 1451–1459, doi:10.4049/jimmunol.177.3.1451.
- Denny, M.F.; Yalavarthi, S.; Zhao, W.; Thacker, S.G.; Anderson, M.; Sandy, A.R.; McCune, W.J.; Kaplan, M.J. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J. Immunol. 2010, 184, 3284–3297, doi:10.4049/jimmunol.0902199.
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498, doi:10.1016/j.cell.2009.09.033.
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 2011, 331, 337–341, doi:10.1126/science.1198469.
- Rosser, E.C.; Oleinika, K.; Tonon, S.; Doyle, R.; Bosma, A.; Carter, N.A.; Harris, K.A.; Jones, S.A.; Klein, N.; Mauri, C. Regulatory B cells are induced by gut microbiota-driven interleukin-1beta and interleukin-6 production. Nat. Med. 2014, 20, 1334–1339, doi:10.1038/nm.3680.
- Mu, Q.; Tavella, V.J.; Kirby, J.L.; Cecere, T.E.; Chung, M.; Lee, J.; Li, S.; Ahmed, S.A.; Eden, K.; Allen, I.C.; et al. Antibiotics ameliorate lupus-like symptoms in mice. Sci. Rep. 2017, 7, 13675, doi:10.1038/s41598-017-14223-0.
- Zhang, H.; Liao, X.; Sparks, J.B.; Luo, X.M. Dynamics of gut microbiota in autoimmune lupus. Appl. Environ. Microbiol. 2014, 80, 7551–7560, doi:10.1128/AEM.02676-14.
- Mu, Q.; Zhang, H.; Liao, X.; Lin, K.; Liu, H.; Edwards, M.R.; Ahmed, S.A.; Yuan, R.; Li, L.; Cecere, T.E.; et al. Control of lupus nephritis by changes of gut microbiota. Microbiome 2017, 5, 73, doi:10.1186/s40168-017-0300-8.
- Andrews, B.S.; Eisenberg, R.A.; Theofilopoulos, A.; Izui, S.; Wilson, C.B.; McConahey, P.; Murphy, E.D.; Roths, J.B.; Dixon, F. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J. Exp. Med. 1978, 148, 1198–1215, doi:10.1084/jem.148.5.1198.
- Rodríguez-Carrio, J.; López, P.; Sánchez, B.; González, S.; Gueimonde, M.; Margolles, A.; de Los Reyes-Gavilán, C.G.; Suárez, A. Intestinal dysbiosis is associated with altered short-chain fatty acids and serum-free fatty acids in systemic lupus erythematosus. Front. Immunol. 2017, 8, 23.
- Zhang, Q.; Yin, X.; Wang, H.; Wu, X.; Li, X.; Li, Y.; Zhang, X.; Fu, C.; Li, H.; Qiu, Y. Fecal Metabolomics and Potential Biomarkers for Systemic Lupus Erythematosus. Front. Immunol. 2019, 10, 976, doi:10.3389/fimmu.2019.00976.
- Zhang, T.; Mohan, C. Caution in studying and interpreting the lupus metabolome. Arthritis Res. Ther. 2020, 22, 172, doi:10.1186/s13075-020-02264-2.
- Eichhorst, A.; Schäfer, A.-L.; Voll, R.E.; Chevalier, N. P55 Influence of dietary fibre and short-chain fatty acids on the pathogenesis of systemic lupus erythematosus. Lupus Science & Medicine 2020; 7, 55, doi:10.1136/lupus-2020-eurolupus.102.
- Gu, J.; Mao, B.; Cui, S.; Liu, X.; Zhang, H.; Zhao, J.; Chen, W. Metagenomic insights into the effects of fructooligosaccharides (FOS) on the composition of luminal and mucosal microbiota in C57BL/6J mice, especially the Bifidobacterium composition. Nutrients 2019, 11, 2431.
- Kang, S.; You, H.J.; Lee, Y.-G.; Jeong, Y.; Johnston, T.V.; Baek, N.-I.; Ku, S.; Ji, G.E. Production, Structural Characterization, and In Vitro Assessment of the Prebiotic Potential of Butyl-Fructooligosaccharides. Int. J. Mol. Sci. 2020, 21, 445.
- Kusumo, P.D.; Maulahela, H.; Utari, A.P.; Surono, I.S.; Soebandrio, A.; Abdullah, M. Probiotic Lactobacillus plantarum IS 10506 supplementation increase SCFA of women with functional constipation. Iran. J. Microbiol. 2019, 11, 389.
- Bhutia, Y.D.; Ganapathy, V. Short, but smart: SCFAs train T cells in the gut to fight autoimmunity in the brain. Immunity 2015, 43, 629–631.
