In eukaryotes, microRNAs (miRNAs) have roles in development, homeostasis, disease and the immune response. Recent work has shown that plant and mammalian miRNAs also mediate cross-kingdom and cross-domain communications, but these studies remain controversial and are lacking critical mechanistic explanations. However, there are several conserved features and homologous components in the distinct Regulatory RNA pathways in eukaryotes and bacteria that could explain how eukaryotic miRNAs could function in such cross-domain communications.
- RNA
- miRNA
- microbiota
- commuincation
- extracellular vesicles
- hfq
- msRNA
- ELNs
- asRNA
Please note: Below is an entry draft based on your previous paper, which is wrirren tightly around the entry title. Since it may not be very comprehensive, we kindly invite you to modify it (both title and content can be replaced) according to your extensive expertise. We believe this entry would be beneficial to generate more views for your work. In addition, no worry about the entry format, we will correct it and add references after the entry is online (you can also send a word file to us, and we will help you with submitting).
1. Introduction
The expansive non-coding regions of eukaryotic genomes are now known to encode many important regulatory elements such as promoters, enhancers and long non-coding RNAs (lncRNAs) that influence the transcription of both protein-coding and non-coding regions [1,2][1][2]. Some regions function as post-transcriptional gene expression regulators, such as microRNAs (miRNAs), small interfering RNAs (siRNAs) and P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) that bind to their targets through complementary base pairing (Table 1).
Illustrating the similarities between eukaryotic and bacterial antisense RNA-mediated regulatory pathways.
|
Eukaryotic RNA |
Length |
Functions |
Mechanisms |
Distribution |
Bacterial RNA |
Length |
Functions |
Mechanisms |
Distribution |
|||||||||||||||||||||||||
|
miRNA |
21-25 nucleotides |
• Translational repression and transcriptional decay • Defence against exogenous viruses • Transposon defence • Epigenetic regulation |
Imperfect complementarity to target RNase III enzymes for biogenesis (Dicer and Drosha) Typically bind the 3’'UTR of target mRNA
In eukaryotes, miRNAs and siRNAs are derived from longer precursor molecules and processed into smaller molecules of approximately 21 to 25 nucleotides in length [3]. Both miRNA and siRNA mediate post-transcriptional gene silencing through the RNA-induced silencing (RISC) complex and the AGO subfamily of Argonaute proteins. The key difference is that miRNAs can bind to targets with imperfect complementarity, enabling one miRNA to target hundreds of messenger RNAs (mRNAs), whereas siRNAs generally require perfect complementarity to their target sequence [4]. Additionally, the siRNA silencing mechanism can be amplified by the action of RNA-dependent RNA polymerases (RdRPs). These enzymes produce secondary siRNAs by using the target mRNA as a template [5,6][5][6]. However, it is unclear whether such amplification occurs in vertebrates, as they do not possess RdRP genes. miRNAs and siRNAs associated with AGO and RISC can act in the defence against exogenous viruses by targeting key viral genes [7,8][7][8]. Small RNAs also prevent transposon transcription through the direction of epigenetic modifications and heterochromatin formation [9,10][9][10]. In particular, the piRNA family of small non-coding RNAs, are a primary form of defence against transposons and are also critical for fertility in animals [10,11][10][11]. piRNAs mediate transcriptional silencing through the PIWI subfamily of Argonaute proteins. Silencing occurs via the ping-pong cycle, during which a piRNA-bound transposon is cleaved by PIWI, generating another piRNA of the opposite orientation [12]. piRNAs are thought to mediate transposon silencing through heterochromatin formation and de novo DNA methylation, although the precise mechanisms through which this occurs are not yet clear [13]. These classes of regulatory RNA are each reminiscent of the distinct classes of bacterial regulatory RNAs. However, evidence suggests that miRNAs specifically are the key mediators of inter-domain communications [14,15,16][14][15][16]. 1.1. miRNA BiogenesismiRNAs were first discovered in 1993 in Caenorhabditis elegans [17]. They are post-transcriptional gene regulators, typically 21–25 nucleotides in length, and are found in almost all plants and animals [18,19,20][18][19][20]. They are transcribed in the nucleus by RNA polymerase II to form a long pre-cursor primary miRNA molecule which creates a hairpin structure (Figure 1A) [21]. In animals, the hairpin is excised by the endonuclease Drosha and a shorter pre-miRNA hairpin molecule forms which is exported into the cytoplasm by exportin-5 [22,23][22][23]. The endonuclease Dicer then cleaves the terminal hairpin leaving an RNA duplex of approximately 22 nucleotides in length. Typically, one of these strands is preferentially loaded onto the RISC complex, directing the enzymatic complex to the target mRNA [24]. The RISC complex consists of the miRNA and an Argonaute protein, although there can also be many other binding partners [25]. The complex can either degrade the target or inhibit its translation, dependent on the level of complementarity [26]. miRNAs can additionally up-regulate the expression of the target mRNA, although this is less common [27]. miRNAs typically bind to the 3′ Untranslated Region (UTR) of the target mRNA with incomplete base pairing, often only requiring complementarity to the “"seed sequence”" of the miRNA, at nucleotides 2–8 [28]. This enables a single miRNA to have many target transcripts. 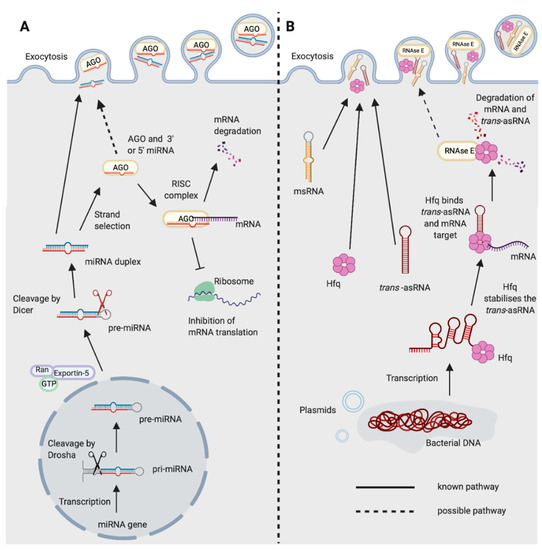 Figure 1. Biogenesis and functions of (A) miRNAs in eukaryotes and (B) trans-asRNAs in bacteria. (A) Eukaryotic miRNAs are transcribed in the nucleus into a pre-cursor primary-miRNA (pri-miRNA) transcript. The pri-miRNA is cleaved by Drosha (black scissors) to form a shorter hairpin, the pre-miRNA. The pre-miRNA is exported from the nucleus by exportin-5 in conjunction with ran-GTP. In the cytoplasm, the pre-miRNA is cleaved further by Dicer (red scissors) to form a miRNA duplex. One strand from the miRNA duplex is preferentially loaded onto Argonaute (AGO) forming the RNA-induced Silencing Complex (RISC). The miRNA of the RISC complex facilitates binding to the target mRNA (requiring only partial complementarity). Catalytically active AGOs (AGO2 in humans) can cleave and degrade the mRNA target. The RISC complex can also inhibit ribosomal translation of mRNAs. Exocytosis of miRNAs in Extracellular Vesicles (EVs) has been shown in eukaryotes [14]. It is not yet known whether the miRNA could be loaded onto AGO prior to exocytosis. (B) Newly transcribed bacterial trans-asRNAs can be stabilised by Hfq. Hfq can also facilitate binding between the trans-asRNA and the mRNA target [29,30][29][30]. In Gram-negative bacteria, Hfq can facilitate binding of the trans-asRNA and mRNA to RNase E which degrades both the asRNA and mRNA [31]. It is unclear whether RNase E can be exported from bacteria, but it does possess a domain capable of binding the membrane of phospholipid vesicles in vitro [32]. Bacteria have been shown to export microRNA-size RNA (msRNA) and trans-asRNAs in EVs [33,34,35][33][34][35]. Hfq can also be exported from bacteria in EVs [36] (Created with BioRender.com). 1.2. miRNAs in DiseaseCurrently, there are 1917 identified miRNA loci in the human genome, with 60% of human protein-coding genes estimated to be regulated by at least one miRNA [37,38][37][38]. They form complex signalling networks which regulate important biological processes such as cell differentiation and proliferation, apoptosis, metabolism, the immune response, development and ageing [39,40,41,42,43][39][40][41][42][43]. Due to the critical roles of miRNAs in such a range of biological processes, they are also ideal targets for pathogens that need to alter their environment to establish infection. Host miRNAs play a fundamental role in shaping the innate and adaptive immune responses, and can also directly target pathogens and inhibit their replication [44,45,46][44][45][46]. Conversely, pathogens can also produce miRNAs to disrupt the immune response, facilitating their own survival [7,47][7][47]. Human Immunodeficiency Virus 1 (HIV-1) encodes a pre-miRNA sequence in its genome (HIV1-miR-H1). HIV-1 exploits the human host’'s miRNA processing machinery to convert this into a functional mature miRNA. This has critical roles in HIV-1 pathogenesis due to its repression of host anti-viral responses, including the host anti-viral miRNA, hsa-miR-149 [48,49,50][48][49][50]. Other viruses capable of producing miRNAs include Herpes Simplex 1, Epstein Barr virus and Cytomegalovirus [51]. Consequently, an increasing number of clinical trials are focusing on the therapeutic value of miRNAs in viral infections. There are also several active clinical trials investigating miRNAs in other human diseases including in autoimmunity, various cancers and cardiovascular disease ([52], and at clinicaltrials.gov). 1.3. miRNAs and BacteriaThe vast majority of studies concerning miRNAs have focused on the role of miRNAs in eukaryotes, or in viruses that exploit eukaryotic cell machinery [7,51,52][7][51][52]. However, an expanding area of research is now showing that miRNAs can alter the composition of the bacteria in the intestinal microbiota of mammalian hosts [14,16][14][16]. Dysbiosis of the intestinal microbiota has been demonstrated in an increasing number of diseases ranging from Inflammatory Bowel Disease and Multiple Sclerosis, to psychological disorders including Bipolar Disorder, Depression and Anxiety amongst many others [53,54,55,56][53][54][55][56]. The mechanisms by which these unfavourable changes occur are largely unclear. Whether miRNAs could play a role in interactions between the more distantly related eukaryotes and bacteria is an emerging concept that requires more thorough consideration. 2. Regulatory RNAs in BacteriaBacteria possess three key classes of regulatory molecules—(i) cis-acting 5′ element non-coding RNAs, (ii) trans-acting small non-coding asRNAs (trans-asRNAs), and (iii) cis-encoded antisense RNAs (cis-asRNAs). Cis-acting 5′ element non-coding RNAs are typically present in the 5′UTR of the mRNA they regulate. Ligand binding induces structural changes in the non-coding RNA, that subsequently influences the transcription of the downstream gene. This type of RNA-mediated regulation includes riboswitches, thermoregulators and pH sensors [57]. Whereas, the bacterial trans-asRNAs and cis-asRNAs recognise their target sequence via complementary base pairing, akin to eukaryotic miRNAs, siRNAs and piRNAs. Eukaryotes and prokaryotes have utilised RNA-dependent mechanisms for various common functions including post-transcriptional gene regulation, as a defence against exogenous viral infections and as a defence against transposons (Table 1). 3. Shared Features of Eukaryotic and Bacterial Regulatory RNA Pathways3.1. A Conserved Protein Fold between AGO, PIWI and CasThe viral-derived RNase H-like protein superfamily is thought to have been a key driver of the evolution of complex life forms on the early earth, as it includes many essential enzymes in both eukaryotes and bacteria [88][58]. Members of the RNase H-like protein superfamily are mediators of many important processes including DNA replication, transposition, gene splicing, and the generation of adaptive immune responses [89][59]. The RNA interference (RNAi) pathways of eukaryotes and the CRISPR anti-viral defence of bacteria both rely on RNase H-like endonucleases. In RNAi, this is the Argonaute and PIWI-like proteins, and in CRISPR, the Cas proteins [89][59]. These enzymes all possess an RNase H fold, one of the most ancient and abundant protein folds known [90][60]. At a glance, these distinct pathways and mechanisms can initially appear to be completely unrelated. However, there are many shared features of these pathways. 3.2. CRISPR RNA and piRNA-Guided ImmunityOne of the most striking similarities between eukaryotic and prokaryotic regulatory RNA pathways is the crRNA-guided CRISPR immunity of prokaryotes and piRNA-guided immunity in metazoans. The piRNAs and crRNAs of these pathways each represent a form of genome-encoded trans-generational adaptive immunity. In prokaryotes, CRISPR mediates defence against invasive nucleic acids from bacteriophage and plasmids [67][61]. In metazoans, piRNA-mediated immunity is the primary defence mechanism against transposable elements in the germline [91][62]. The majority of piRNAs are derived from piRNA clusters [92][63]. Similarly, crRNAs are also derived from distinct regions in the prokaryotic genome, the CRISPR loci [67][61]. In prokaryotes, short sequences of the invasive DNA are integrated into the CRISPR loci by the Cas endonuclease, separated by repeat sequences. In eukaryotes, both invasive RNA and DNA viruses can be integrated into piRNA clusters [93,94][64][65]. Both piRNAs and crRNAs are classes of regulatory RNAs that are complementary in sequence to the invasive nucleic acid, and that guide an endonuclease complex to the foreign target sequence [94][65]. In CRISPR, this is the Cas endonuclease that is guided by crRNAs, whereas piRNAs guide the PIWI endonuclease to the target sequence. In contrast to CRISPR, piRNAs can also direct histone modifications that mediate gene silencing [95][66]. These pathways are prime examples of the parallel functions and mechanisms of the distinct regulatory RNA pathways in eukaryotes and bacteria. 3.3. Shared Features of mRNA Decay in Eukaryotes and BacteriaIn eukaryotes, newly synthesised mRNAs are characterised by a 5′ 7-methylguanylate (m7G) cap and 3′ poly-adenylation [96][67]. General mRNA decay occurs due to decapping of the mRNAs and removal of the poly (A) tails in cytoplasmic Processing bodies (P-bodies) [97][68]. In bacteria, the degradosome is the key mediator of bulk RNA turnover, and newly synthesised bacterial mRNAs possess a 5′ triphosphate group that must be converted to a 5′ monophosphate group for efficient recognition by RNase E or RNase Y of the degradosome [98][69]. Interestingly, both miRNAs and siRNAs in eukaryotes are also characterised by 5′ monophosphate groups, and the enzyme responsible for this conversion in E. coli, RppH, is closely related to DCP2 of the decapping complex in eukaryotes [99,100,101][70][71][72]. The DCP2 decapping complex is also known to be involved in miRNA-mediated gene silencing in eukaryotes [102][73]. The 5′ monophosphate may enable the association of miRNAs and siRNAs with the nucleases of the bacterial degradosomes. Whether this shared evolutionary relationship between decapping enzymes could also enable miRNA-mediated regulation of bacterial gene expression through the degradosome, is another interesting prospect. A further example of this shared relationship between eukaryotic and bacterial RNA decay pathways is also demonstrated by Hfq of the trans-asRNA pathway, and its likeness to the eukaryotic Sm proteins. 3.4. Bacterial Hfq is Reminiscent of Eukaryotic Decapping ProteinsThe function of Hfq is somewhat analogous to the RISC complex, as it facilitates interactions between trans-asRNAs and their target mRNAs [103][74]. Yet, it bears the most striking structural and functional similarities to the eukaryotic Sm and Sm-like proteins [104,105][75][76]. It contains the conserved Sm1 motif and, similar to the eukaryotic proteins, forms an oligomeric ring-shaped structure. Hfq facilitates the regulation of mRNA stability through interactions with trans-asRNAs, whereas in eukaryotes the Sm-like protein, Lsm1, is involved in mRNA decapping and degradation [106][77]. These conserved structural and functional components may enable trans-asRNAs of bacteria to interact with the mRNA decay machinery in eukaryotes. Alternatively, they may enable miRNAs that function through decapping enzymes to influence bacterial gene expression. Investigating the ability of eukaryotic regulatory RNAs to complex with Hfq or the bacterial degradosome, and of trans-asRNAs to interact with eukaryotic Sm-like proteins could reveal novel mechanisms of host-bacterial interactions. 4. miRNAs Facilitate Inter-Kingdom CommunicationsThere are several examples of inter-kingdom communications mediated by miRNAs and miRNA-like molecules amongst the eukaryotes. The processes of RNA release and the mechanisms by which they remain resistant to degradation in the environment are critical to RNA-mediated inter-kingdom interactions. Extracellular Vesicles (EVs) containing RNA as cargo offer a route by which this communication could be mediated. 5. Mechanisms for RNA-based Communication between Eukaryotes and Bacteria5.1. Bacterial asRNAs Can Hijack Eukaryotic RNAi PathwaysSeveral studies have illustrated the presence of msRNAs in bacterial genomes using bioinformatic approaches over the last decade [125,126,127][78][79][80]. These bacterial-derived msRNAs can be delivered into host cells and incorporate into the eukaryotic host’'s miRNA-processing machinery, thereby serving important functional roles for the bacterium [47,128][47][81]. For example, an msRNA derived from an RNA stem-loop in Mycobacterium marinum associates with eukaryotic RISC [128][81]. Furthermore, Salmonella can release RNA into the cytosol of infected IECs, and this RNA is processed by human AGO2, in a Dicer-independent fashion, into small functional msRNAs of 22 nucleotides in length. One of these miRNA-like molecules, Sal-1, serves as a critical virulence factor, facilitating the intracellular replication and survival of the bacteria [47]. These data demonstrate a role for bacterial-derived RNA molecules in the successful colonisation by human pathogens. Similarly, it was observed that the periodontal pathogens Aggregatibacter actinomycetemcomitans, P. gingivalis, and Treponema denticola produce msRNAs in EVs that could enter eukaryotic fibroblast cells in vitro [129][82]. Exogenous transfection of some of the most abundantly expressed msRNAs by these bacteria led to a decrease in the anti-inflammatory cytokines IL-5 and IL-13 by Jurkat T cells [129][82]. These pathogens have been implicated in periodontitis, which is driven by the inflammatory response to the bacteria of the oral microbiota [130][83]. This work, therefore, highlights bacterial RNAs as a potential novel therapeutic target. Additionally, trans-acting asRNAs OxyS and DsrA from E. coli, can impair C. elegans chemosensory behaviour and longevity through down-regulation of che-2 and the diacylglycerol lipase gene F42G9.6, respectively [35]. This is an RDE-4 dependent process, with RDE-4 being a key component of C. elegans RNAi, that processes dsRNA by complexing with Dicer and subsequently worm argonaute, RDE-1 [35,131][35][84]. In E. coli OxyS and DsrA function to co-ordinate the stress response through the regulation of the protein sigma factor RpoS in an Hfq-dependent manner [132][85]. Together, these studies show that a number of bacteria are known to produce RNAs that could mimic eukaryotic regulatory RNAs and feed into eukaryotic RNAi processing pathways. In addition, bacterial asRNAs can also have additional distinct functions in the eukaryotic host. Whether these relationships extend to host-microbiota interactions is an intriguing prospect that warrants further study. Investigating whether the production of RNA by members of the microbiota can modulate relationships with the host, and vice versa will be a key part of translating correlative microbiota studies into clinical benefits. 5.2. Proposal of Mechanisms for Regulatory RNAs in Inter-domain CommunicationsThis review of the current literature leads us to propose two potential mechanisms by which regulatory RNAs could mediate bi-directional communications between eukaryotes and bacteria. (1) RNA molecules produced by one organism could integrate into one of the distinct regulatory RNA pathways present in the other. For example, the RNAs produced by Salmonella become functional upon processing by eukaryotic RNAi machinery, such as AGO2 and Dicer [47]. This is also true for E. coli, which exploits C. elegans RNAi machinery to regulate eukaryotic gene expression through its own bacterial trans-asRNAs [35] (2) RNAs are exported in EVs in conjunction with the molecules required for their functioning in the recipient (Figure 2). This is shown by Hfq which is exported in EVs by Yersinia pestis, and in an Acinetobacter baumannii Hfq mutant which displays reduced EV production [36,133][36][86]. Furthermore, eukaryotic Argonaute has also been detected in EVs [134][87]. These mechanisms could also occur in combination, with both host and recipient molecules important in inter-domain regulatory RNA-mediated communications. 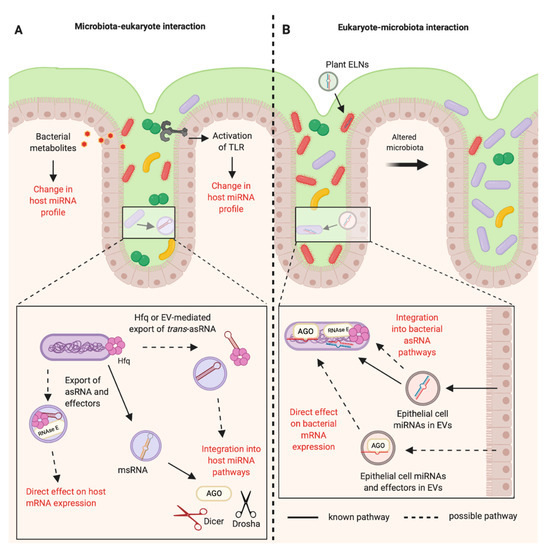 Figure 2. RNA-mediated communications between the mammalian host and the bacteria of the gastrointestinal microbiota. (A) The bacteria of the mammalian intestinal microbiota can indirectly alter the host miRNA profile through the production of metabolites and activation of Toll-like Receptors (TLRs). It is known that bacteria can produce microRNA-size RNAs (msRNAs) in Extracellular Vesicles (EVs) that can integrate into host miRNA pathways [47]. Other RNAs such as trans-asRNAs could be exported in vesicles and could similarly integrate into host pathways. It is also possible that asRNAs could be exported along with their effectors in EVs (such as Hfq and RNase E) and could directly influence host gene expression. EV-independent export of asRNAs could be mediated by Hfq, as Hfq can create holes in the bacterial cell membrane [79]. (B) Plant exosome-like nanoparticles (ELNs) from diet and EVs from intestinal epithelial cells containing miRNAs can enter members of the microbiota and influence bacterial gene expression and growth [14,16][14][16]. These miRNAs could be exported with effectors such as Argonaute (AGO) to directly alter bacterial gene expression. Alternatively, eukaryotic miRNAs could become functional upon integration into existing bacterial asRNA regulatory pathways (Created with BioRender.com). |
References
- Fang, Y.; Fullwood, M. Roles, Functions, and mechanisms of long non-coding RNAs in cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54.
- Maston, G.; Evans, S.; Green, M. Transcriptional regulatory elements in the human genome. Annu. Rev. Genom. Hum. Genet. 2006, 7, 29–59.
- Carthew, R.; Sontheimer, E. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655.
- Lam, J.; Chow, M.; Zhang, Y.; Leung, S. siRNA versus miRNA as therapeutics for gene silencing. Molecular Therapy Nucleic Acids 2015, 4, e252.
- Sijen, T.; Fleenor, J.; Simmer, F.; Thijssen, K.; Parrish, S.; Timmons, L.; Plasterk, R.; Fire, A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 2001, 107, 465–476.
- Zhang, C.; Ruvkun, G. New insights into siRNA amplification and RNAi. RNA Biol. 2012, 9, 1045–1049.
- Kaul, D.; Khanna, A.; Suman Evidence and nature of a novel miRNA encoded by HIV-1. Proc. Indian Acad. Sci. 2006, 72, 91–95.
- Zhou, R.; Rana, T. RNA-based mechanisms regulating host-virus interactions. Immunol. Rev. 2013, 253, 97–111.
- Lippman, Z.; Martienssen, R. The role of RNA interference in heterochromatic silencing. Nature 2004, 431, 364–370.
- Shalgi, R.; Pilpel, Y.; Oren, M. Repression of transposable-elements—A microRNA anti-cancer defense mechanism? Trends Genet. 2010, 26, 253–259.
- Castañeda, J.; Genzor, P.; Bortvin, A. piRNAs, transposon silencing, and germline genome integrity. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2011, 714, 95–104.
- Czech, B.; Hannon, G. One loop to rule them all: The Ping-Pong cycle and piRNA-guided silencing. Trends Biochem. Sci. 2016, 41, 324–337.
- Le Thomas, A.; Rogers, A.; Webster, A.; Marinov, G.; Liao, S.; Perkins, E.; Hur, J.; Aravin, A.; Toth, K. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399.
- Liu, S.; da Cunha, A.; Rezende, R.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.; Gandhi, R.; Weiner, H. The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43.
- Sundaram, K.; Miller, D.; Kumar, A.; Teng, Y.; Sayed, M.; Mu, J.; Lei, C.; Sriwastva, M.; Zhang, L.; Jun, Y.; et al. Plant-derived exosomal nanoparticles inhibit pathogenicity of Porphyromonas gingivalis. iScience 2019, 21, 308–327.
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-derived exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe 2018, 24, 637–652.e8.
- Lee, R.; Feinbaum, R.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Bruscella, P.; Bottini, S.; Baudesson, C.; Pawlotsky, J.; Feray, C.; Trabucchi, M. Viruses and miRNAs: More friends than foes. Front. Microbiol. 2017, 8, 824.
- Lee, H.; Li, L.; Gu, W.; Xue, Z.; Crosthwaite, S.; Pertsemlidis, A.; Lewis, Z.; Freitag, M.; Selker, E.; Mello, C.; et al. Diverse pathways generate MicroRNA-like RNAs and dicer-independent small interfering RNAs in Fungi. Mol. Cell 2010, 38, 803–814.
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Et Biophys. Acta (Bba) Mol. Cell Res. 2010, 1803, 1231–1243.
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419.
- Lee, Y. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670.
- Yi, R.; Qin, Y.; Macara, I.; Cullen, B. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016.
- Kuchenbauer, F.; Mah, S.; Heuser, M.; McPherson, A.; Rüschmann, J.; Rouhi, A.; Berg, T.; Bullinger, L.; Argiropoulos, B.; Morin, R.; et a. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood 2011, 118, 3350–3358.
- Rivas, F.; Tolia, N.; Song, J.; Aragon, J.; Liu, J.; Hannon, G.; Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005, 12, 340–349.
- Zeng, Y.; Yi, R.; Cullen, B. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 9779–9784.
- Vasudevan, S. Posttranscriptional upregulation by MicroRNAs. Wiley Interdiscip. Rev. RNA 2011, 3, 311–330.e
- Lewis, B.; Shih, I.; Jones-Rhoades, M.; Bartel, D.; Burge, C. Prediction of mammalian MicroRNA targets. Cell 2003, 115, 787–798.
- Baek, Y.; Jang, K.; Lee, H.; Yoon, S.; Baek, A.; Lee, K.; Kim, D. The bacterial endoribonuclease Rnase E can cleave RNA in the absence of the RNA chaperone Hfq. J. Biol. Chem. 2019, 294, 16465–16478.
- Ikeda, Y.; Yagi, M.; Morita, T.; Aiba, H. Hfq binding at RhlB-recognition region of Rnase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 2010, 79, 419–432.
- De Lay, N.; Gottesman, S. Rnase E finds some sRNAs Stimulating. Mol. Cell 2012, 47, 825–826.
- Khemici, V.; Poljak, L.; Luisi, B.; Carpousis, A. The Rnase E of Escherichia coli is a membrane-binding protein. Mol. Microbiol. 2008, 70, 799–813.
- Dauros-Singorenko, P.; Blenkiron, C.; Phillips, A.; Swift, S. The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol. Lett. 2018, 365, doi:10.1093/femsle/fny023.
- Ghosal, A.; Upadhyaya, B.; Fritz, J.; Heintz-Buschart, A.; Desai, M.; Yusuf, D.; Huang, D.; Baumuratov, A.; Wang, K.; Galas, D.; et al. The extracellular RNA complement of Escherichia coli. MicrobiologyOpen 2015, 4, 252–266.
- Liu, H.; Wang, X.; Wang, H.; Wu, J.; Ren, J.; Meng, L.; Wu, Q.; Dong, H.; Wu, J.; Kao, T.; et al.. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 2012, 3, 1073.
- Eddy, J.; Gielda, L.; Caulfield, A.; Rangel, S.; Lathem, W. Production of outer membrane vesicles by the plague pathogen Yersinia pestis. PloS ONE 2014, 9, e107002.
- Friedman, R.; Farh, K.; Burge, C.; Bartel, D. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105.
- Mirbase.org. Mirbase. [online]. 2020. Available online: http://www.mirbase.org/summary.shtml?org=hsa (accessed on 10 April 2020).
- Colamatteo, A.; Micillo, T.; Bruzzaniti, S.; Fusco, C.; Garavelli, S.; De Rosa, V.; Galgani, M.; Spagnuolo, M.; Di Rella, F.; Puca, A.; et al. Metabolism and autoimmune responses: The microRNA connection. Front. Immunol. 2019, 10, 1969.
- Hwang, H.; Mendell, J. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2006, 94, 776–780.
- Ivey, K.; Srivastava, D. microRNAs as Developmental Regulators. Cold Spring Harb. Perspect. Biol. 2015, 7, a008144.
- Slattery, M.; Mullany, L.; Sakoda, L.; Wolff, R.; Samowitz, W.; Herrick, J. Dysregulated genes and miRNAs in the apoptosis pathway in colorectal cancer patients. Apoptosis 2018, 23, 237–250.
- Smith-Vikos, T.; Slack, F. MicroRNAs and their roles in aging. J. Cell Sci. 2012, 125, 7–17.
- Bai, X.; Nicot, C. miR-28-3p Is a cellular restriction factor that inhibits human T cell leukemia virus, type 1 (HTLV-1) replication and virus infection. J. Biol. Chem. 2015, 290, 5381–5390.
- He, J.; Ji, Y.; Li, A.; Zhang, Q.; Song, W.; Li, Y.; Huang, H.; Qian, J.; Zhai, A.; Yu, X.; et al. MiR-122 directly inhibits human papillomavirus E6 gene and enhances interferon signaling through blocking suppressor of cytokine signaling 1 in SiHa cells. PloS ONE 2014, 9, e108410.
- Zhou, X.; Li, X.; Wu, M. miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct. Target. Ther. 2018, 3, 1–13.
- Gu, H.; Zhao, C.; Zhang, T.; Liang, H.; Wang, X.; Pan, Y.; Chen, X.; Zhao, Q.; Li, D.; Liu, F.; et al. Salmonella produce microRNA-like RNA fragment Sal-1 in the infected cells to facilitate intracellular survival. Sci. Rep. 2017, 7, 1–12.
- Hariharan, M.; Scaria, V.; Pillai, B.; Brahmachari, S. Targets for human encoded microRNAs in HIV genes. Biochem. Biophys. Res. Commun. 2005, 337, 1214–1218.
- Kaul, D.; Ahlawat, A.; Gupta, S. HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses to infection. Mol. Cell. Biochem. 2008, 323, 143–148.
- Kogan, M.; Rappaport, J. HIV-1 accessory protein Vpr: Relevance in the pathogenesis of HIV and potential for therapeutic intervention. Retrovirology 2011, 8, 25.
- Piedade, D.; Azevedo-Pereira, J. The role of microRNAs in the pathogenesis of herpes virus infection. Viruses 2016, 8, 156.
- Hanna, J.; Hossain, G.; Kocerha, J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019, 10, 478.
- De Palma, G.; Lynch, M.; Lu, J.; Dang, V.; Deng, Y.; Jury, J.; Umeh, G.; Miranda, P.; Pigrau Pastor, M.; Sidani, S.; et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 2017, 9, eaaf6397.
- Evans, S.; Bassis, C.; Hein, R.; Assari, S.; Flowers, S.; Kelly, M.; Young, V.; Ellingrod, V.; McInnis, M. The gut microbiome composition associates with bipolar disorder and illness severity. J. Psychiatr. Res. 2017, 87, 23–29.
- Pinto-Sanchez, M.; Hall, G.; Ghajar, K.; Nardelli, A.; Bolino, C.; Lau, J.; Martin, F.; Cominetti, O.; Welsh, C.; Rieder, A.; et al. Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: A pilot study in patients with irritable bowel syndrome. Gastroenterology 2017, 153, 448–459.e8.
- Zheng, P.; Zeng, B.; Zhou, C.; Liu, M.; Fang, Z.; Xu, X.; Zeng, L.; Chen, J.; Fan, S.; Du, X.; et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 2016, 21, 786–796.
- Saberi, F.; Kamali, M.; Najafi, A.; Yazdanparast, A.; Moghaddam, M. Natural antisense RNAs as mRNA regulatory elements in bacteria: A review on function and applications. Cell. Mol. Biol. Lett. 2016, 21, 6.
- Moelling, K.; Broecker, F. The reverse transcriptase-RNase H: From viruses to antiviral defense. Ann. N. Y. Acad. Sci. 2015, 1341, 126–135.
- Moelling, K.; Broecker, F.; Russo, G.; Sunagawa, S. RNase H as gene modifier, driver of evolution and antiviral defense. Front. Microbiol. 2017, 8, 1745.
- Majorek, K.; Dunin-Horkawicz, S.; Steczkiewicz, K.; Muszewska, A.; Nowotny, M.; Ginalski, K.; Bujnicki, J. The RNase H-like superfamily: New members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 2014, 42, 4160–4179.
- Rath, D.; Amlinger, L.; Rath, A.; Lundgren, M. The CRISPR-Cas immune system: Biology, mechanisms and applications. Biochimie 2015, 117, 119–128.
- Tóth, K.; Pezic, D.; Stuwe, E.; Webster, A. The piRNA pathway guards the germline genome against transposable elements. Non-Coding RNA Reprod. Syst. 2015, 886, 51–77.
- Yamanaka, S.; Siomi, M.; Siomi, H. piRNA clusters and open chromatin structure. Mob. DNA 2014, 5, 22.
- Horie, M.; Honda, T.; Suzuki, Y.; Kobayashi, Y.; Daito, T.; Oshida, T.; Ikuta, K.; Jern, P.; Gojobori, T.; Coffin, J.; et al. Endogenous non-retroviral RNA virus elements in mammalian genomes. Nature 2010, 463, 84–87.
- Ophinni, Y.; Palatini, U.; Hayashi, Y.; Parrish, N. piRNA-guided CRISPR-like immunity in eukaryotes. Trends Immunol. 2019, 40, 998–1010.
- Aravin, A.; Bourc’his, D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008, 22, 970–975.
- Gallie, D. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991, 5, 2108–2116.
- Kushner, S. mRNA Decay in prokaryotes and eukaryotes: Different approaches to a similar problem. Int. Union Biochem. Mol. Biol. Life 2004, 56, 585–594.
- Laalami, S.; Zig, L.; Putzer, H. Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 2013, 71, 1799–1828.
- Deana, A.; Celesnik, H.; Belasco, J. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 2008, 451, 355–358.
- Dunckley, T.; Parker, R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999, 18, 5411–5422.
- Wang, Q.; Zhang, D.; Guan, Z.; Li, D.; Pei, K.; Liu, J.; Zou, T.; Yin, P. DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia Coli. Nucleic Acids Res. 2018, 46, 6880–6892.
- Rehwinkel, J.; Behm-Ansmant, I.; Gatfield, D.; Izaurralde, E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 2005, 11, 1640–1647.
- Hoekzema, M.; Romilly, C.; Holmqvist, E.; Wagner, E. Hfq-dependent mRNA unfolding promotes sRNA -based inhibition of translation. EMBO J. 2019, 38, e101199.
- Møller, T.; Franch, T.; Højrup, P.; Keene, D.; Bächinger, H.; Brennan, R.; Valentin-Hansen, P. Hfq: A bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 2002, 9, 23–30.
- Sauter, C.; Basquin, J.; Suck, D. Sm-like proteins in Eubacteria: The crystal structure of the Hfq protein from Escherichia Coli. Nucleic Acids Res. 2003, 31, 4091–4098.
- Chowdhury, A.; Tharun, S. Activation of decapping involves binding of the mRNA and facilitation of the post-binding steps by the Lsm1-7-Pat1 complex. RNA 2009, 15, 1837–1848.
- Dang, T.; Tyagi, S.; D’Cunha, G.; Bhave, M.; Crawford, R.; Ivanova, E. Computational prediction of microRNAs in marine bacteria of the genus Thalassospira. PloS ONE 2019, 14, e0212996.
- Kang, S.; Choi, J.; Lee, Y.; Hong, S.; Lee, H. Identification of microRNA-size, small RNAs in Escherichia coli. Curr. Microbiol. 2013, 67, 609–613.
- Lee, H.; Hong, S. Analysis of microRNA-size, small RNAs in Streptococcus mutans by deep sequencing. Fems Microbiol. Lett. 2011, 326, 131–136.
- Furuse, Y.; Finethy, R.; Saka, H.; Xet-Mull, A.; Sisk, D.; Smith, K.; Lee, S.; Coers, J.; Valdivia, R.; Tobin, D.; et al. Search for MicroRNAs expressed by intracellular bacterial pathogens in infected mammalian cells. PloS ONE 2014, 9, e106434.
- Choi, J.; Kim, S.; Hong, S.; Lee, H. Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J. Dent. Res. 2017, 96, 458–466.
- Cekici, A.; Kantarci, A.; Hasturk, H.; Van Dyke, T. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontology 2000 2013, 64, 57–80.
- Raman, P.; Zaghab, S.; Traver, E.; Jose, A. The double-stranded RNA binding protein RDE-4 can act cell autonomously during feeding RNAi in C. elegans. Nucleic Acids Res. 2017, 45, 8463–8473.
- Liu, J.; Bittker, J.; Lonshteyn, M.; Liu, D. Functional dissection of sRNA translational regulators by nonhomologous random recombination and In Vivo selection. Chem. Biol. 2005, 12, 757–767.
- Kuo, H.; Chao, H.; Liao, P.; Hsu, L.; Chang, K.; Tung, C.; Chen, C.; Liou, M. Functional characterization of Acinetobacter baumannii lacking the RNA chaperone Hfq. Front. Microbiol. 2017, 8, 2068.
- Chow, F.; Koutsovoulos, G.; Ovando-Vázquez, C.; Neophytou, K.; Bermúdez-Barrientos, J.; Laetsch, D.; Robertson, E.; Kumar, S.; Claycomb, J.; Blaxter, M.; et al. Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res. 2019, 47, 3594–3606.
