In recent years, there has been an increase in knowledge of cancer, accompanied by a technological development that gives rise to medical oncology. An instrument that allows the implementation of individualized therapeutic strategies is the liquid biopsy. Currently, it is the most innovative methodology in medical oncology. Its high potential as a tool for screening and early detection, the possibility of assessing the patient’s condition after diagnosis and relapse, as well as the effectiveness of real-time treatments in different types of cancer. Liquid biopsy is capable of overcoming the limitations of tissue biopsies. The elements that compose the liquid biopsy are circulating tumor cells, circulating tumor nucleic acids, free of cells or contained in exosomes, microvesicle and platelets. Liquid biopsy studies are performed on various biofluids extracted in a non-invasive way, and they can be performed both from the blood and in urine, saliva or cerebrospinal fluid. The development of genotyping techniques, using the elements that make up liquid biopsy, make it possible to detect mutations, intertumoral and intratumoral heterogeneity, and provide molecular information on cancer for application in medical oncology in an individualized way in different types of tumors. Therefore, liquid biopsy has the potential to change the way medical oncology could predict the course of the disease.
- liquid biopsy
- biofluids
- biomarkers
1. Introduction
The selection of biofluid (Figure 1) to obtain information about the disease will depend on the tumor and the accessibility of the sample. In the biofluids, there is molecular information provided by the genetic and epigenetic landscape, systematically following the tumor genomic evolution [[1]].
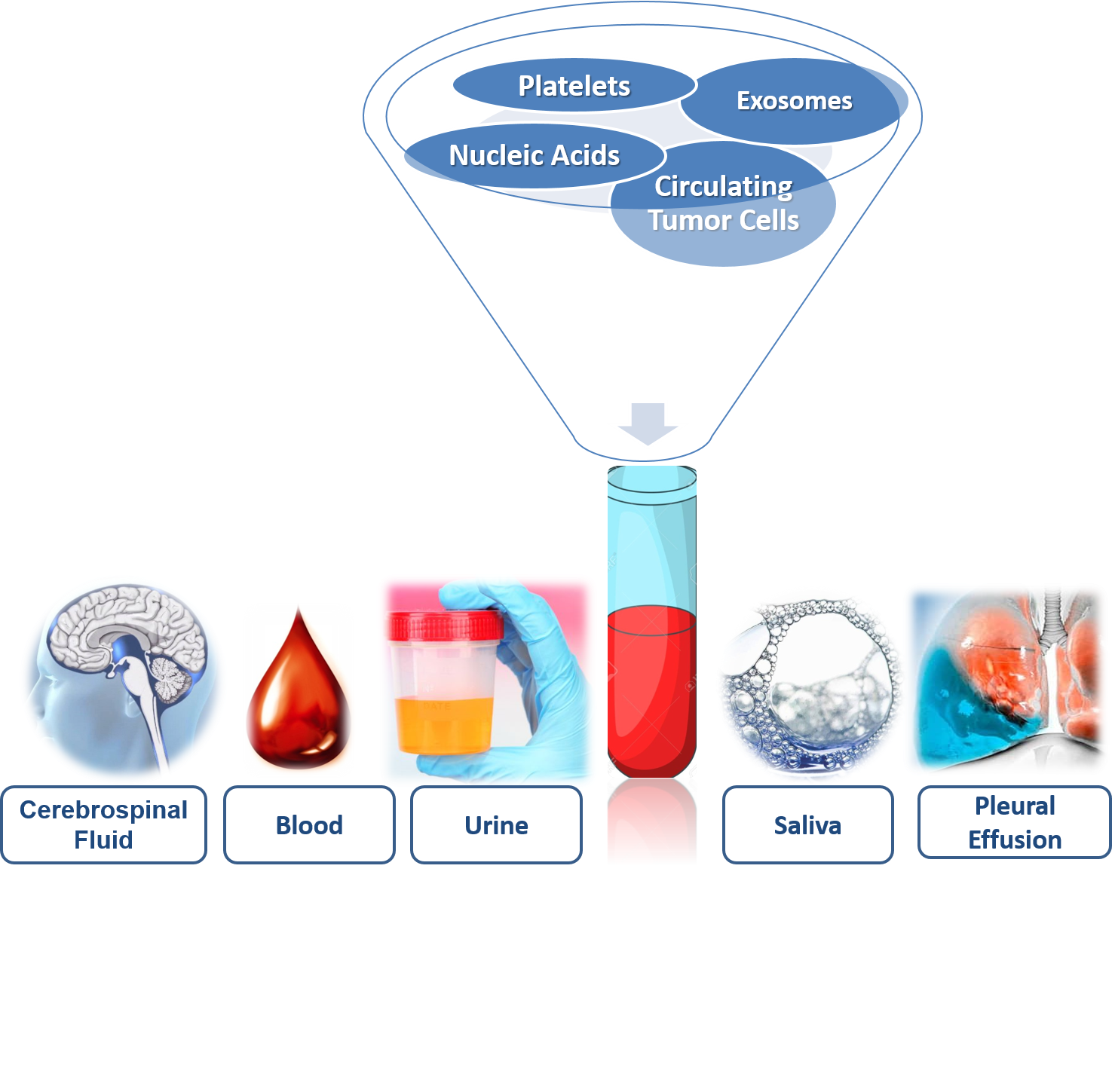
2. Biofluids: Storage of Biomarkers
In malignancies with metastatic capacity such as breast, lung, colon or prostate cancers, once the primitive tumor has invaded the local extracellular matrices, its tumor cells will be able to migrate to distant locations in the body and establish secondary outbreaks, following different dissemination routes: direct, lymphatic and hematogenous. For this reason, CTCs and nucleic acids from the primary tumor can be found in the blood, thus blood is the most commonly used biofluid in the search for tumor biomarkers [[2]]. The obtaining of the sample, by means of blood drawn (simple and low-invasive technique), provides dynamic information on the progress and evolution of the type of cancer.
In the plasma and/or serum, there are tumor marker proteins such as the carcinoembryonic antigen (CEA), the carbohydrate antigen 19-9 (CA19.9) or the prostate-specific antigen (PSA). In addition, the current revolution in the field of blood biomarkers makes it possible to study circulating nucleic acids, both circulating tumor DNA (ctDNA) and circulating tumor RNA (ctRNA), using highly sensitive genomic techniques [[3]].
Urine is divided into sediments, which allows the macroscopic study of crystalline structures in the form of salts and supernatants, where we find proteins, metabolites, nucleic acids and vesicles of extracellular origin [[4]]. The prostate-specific antigen (PSA) is found to increase in the urine in patients with prostate cancer, although the clinical routine determines it in blood [[4]]. Urine DNA comes from glomerular filtration where the fragments are 100 base pairs, although it depends on the patient’s condition. The enzymatic activity of DNAse-1 produces a high fragmentation of DNA with high (≥1 kpb) or low (<100 bp) molecular weight [[5]]. Urine DNA analysis has determined mutations in KRAS in pancreatic and colorectal cancers. In addition, in non-small-cell lung cancer, the epidermal growth factor receptor (EGFR) has detected mutations with sensitivities similar to those obtained in plasma. However, the detection of mRNA is not possible due to the action of RNases, although it is possible to detect micro RNA found inside urine exosomes [[1]].
In the saliva (hypotonic solution), there exist isolated proteins, DNA and RNA, and metabolites and microbiota, that are also present in the blood. Thus, their concentration changes can be used as biomarkers to detect early-stage cancer or to monitor the response to therapeutic management [[6]]. Salivary diagnostics are non-invasive, easy to use tools for patient specimen collection. Saliva testing potentially allows the patient to gather their own saliva samples, even at home, thus saving healthcare costs, enabling convenient and multiple sampling as well as having a positive impact on patient compliance [[7]].
In pancreatic cancer, the salivary messenger RNA can be used to detect KRAS, Methyl-CpG Binding Domain Protein 3-Like 2 (MBD3L2), Acrosomal Vesicle Protein 1 (ACRV1) and Dolichyl-Phosphate Mannosyltransferase Subunit 1 (DPM1) with 90% sensitivity and 95% specificity. All biomarkers together had greater diagnostic power than any single biomarker [[8]]. In addition, miRNA showed their importance in salivary diagnosis because they were expressed differentially in saliva samples from pancreatic cancer patients compared with controls [[9][10]]. Lau et al. [[11]] provide the role of exosomes derived from pancreatic cancer in the development of salivary biomarkers. In lung cancer, the salivary transcriptome was analyzed using mRNA as a biomarker and could differentiate lung cancer patients from control subjects [[12][13]]. In addition, some proteins such as haptoglobin hp2 (HP), zinc 2-glycoprotein (AZGP1) and calprotectin were differentially identified with high levels of sensitivity and specificity in the saliva of lung cancer patients [[12][13]]. Breast cancer expressed different biomarkers, similar to those expressed in saliva: epidermal growth factor (EGF), p53, human epidermal growth factor receptor 2 (HER2/neu) and carbohydrate antigen 15-3 (CA 15-3) [[6]]. Salivary proteins for the detection of gastric cancer were detected as biomarkers, discriminating between healthy controls and patients [[14]]. Finally, the quantity and quality of salivary DNA is similar to plasma and has been used to detect mutations of phosphatidylinositol 3-kinase (PI3K), the cyclin-dependent inhibitor of kinase 2A (CDKN2A), F-box and WD repeat domain-containing 7 (FBXW7), HRAS and KRAS, with a 100% detection sensitivity with tumors located in the oral cavity [[15]].
Although obtaining CSF involves an invasive procedure, it is a better alternative in tumors located in the central nervous system where most of the patients analyzed had circulating nucleic acids in CSF but not in the plasma [[2]]. The purpose of this blood test is for enumerating CTCs. CTCs are cancer cells that detach from a primary tumor and travel through the bloodstream or lymphatic system to other parts of the body an LB that can be used at any time during a patient’s course of the disease [[1][2]].
References
- Giulia Siravegna; Silvia Marsoni; S. Siena; Alberto Bardelli; Integrating liquid biopsies into the management of cancer. Nature Reviews Clinical Oncology 2017, 14, 531-548, 10.1038/nrclinonc.2017.14.
- Klaus Pantel; Catherine Alix-Panabières; Circulating tumour cells in cancer patients: challenges and perspectives. Trends in Molecular Medicine 2010, 16, 398-406, 10.1016/j.molmed.2010.07.001.
- Alberto Bardelli; Klaus Pantel; Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172-179, 10.1016/j.ccell.2017.01.002.
- Guan Hee Tan; Gregory Nason; Khaled Ajib; Dixon Teck Sing Woon; Jaime Herrera-Caceres; Omar Alhunaidi; Nathan Perlis; Smarter screening for prostate cancer. World Journal of Urology 2019, 37, 991-999, 10.1007/s00345-019-02719-5.
- Peter B Gahan; M. Stroun; The Biology of Circulating Nucleic Acids in Plasma and Serum (CNAPS). Recoding: Expansion of Decoding Rules Enriches Gene Expression 2010, 25, 167-189, 10.1007/978-3-642-12617-8_10.
- Elisa Cançado Porto-Mascarenhas; Daniele Xavier Assad; Hélène Chardin; David Gozal; Graziela De Luca Canto; Ana Carolina Acevedo; Eliete Neves Silva Guerra; Salivary biomarkers in the diagnosis of breast cancer: A review. Critical Reviews in Oncology/Hematology 2017, 110, 62-73, 10.1016/j.critrevonc.2016.12.009.
- Herenia P Lawrence; Salivary markers of systemic disease: noninvasive diagnosis of disease and monitoring of general health.. Journal (Canadian Dental Association) 2002, 68, 170-174.
- Lei Zhang; James J. Farrell; Hui Zhou; David Elashoff; David Akin; No–Hee Park; David Chia; David T. Wong; Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer.. Gastroenterology 2009, 138, 949-957, 10.1053/j.gastro.2009.11.010.
- Song Gao; Lian-Yu Chen; Peng Wang; Lu-Ming Liu; Zhen Chen; MicroRNA Expression in Salivary Supernatant of Patients with Pancreatic Cancer and Its Relationship with ZHENG. BioMed Research International 2014, 2014, 1-8, 10.1155/2014/756347.
- Marine Humeau; Alix Vignolle-Vidoni; Flavie Sicard; Frederic Martins; Barbara Bournet; Louis Buscail; Jérôme Torrisani; Pierre Cordelier; Salivary MicroRNA in Pancreatic Cancer Patients. PLOS ONE 2015, 10, e0130996, 10.1371/journal.pone.0130996.
- Chang Lau; Yong Kim; David Chia; Nadine Spielmann; Guido Eibl; David Elashoff; Fang Wei; Yi-Ling Lin; Aune Moro; Tristan Grogan; et al.Samantha ChiangEric FeinsteinChristopher SchaferJames FarrellDavid T. W. Wong Role of Pancreatic Cancer-derived Exosomes in Salivary Biomarker Development*. Journal of Biological Chemistry 2013, 288, 26888-26897, 10.1074/jbc.M113.452458.
- Xiaozhou Li; TianYue Yang; Junxiu Lin; Spectral analysis of human saliva for detection of lung cancer using surface-enhanced Raman spectroscopy. Journal of Biomedical Optics 2012, 17, 37003, 10.1117/1.jbo.17.3.037003.
- Lei Zhang; Hua Xiao; Hui Zhou; Silverio Santiago; Jay M. Lee; Edward B. Garon; Jieping Yang; Ole Brinkmann; Xinmin Yan; David Akin; David Chia; David Elashoff; No-Hee Park; David T. W. Wong; Development of transcriptomic biomarker signature in human saliva to detect lung cancer.. Cellular and Molecular Life Sciences 2012, 69, 3341-3350, 10.1007/s00018-012-1027-0.
- Zheng-Zhi Wu; Ji-Guo Wang; Xiao-Li Zhang; Diagnostic model of saliva protein finger print analysis of patients with gastric cancer.. World Journal of Gastroenterology 2009, 15, 865–870.
- Xiaoqian Wang; Karolina Elżbieta Kaczor-Urbanowicz; David T. W. Wong; Salivary biomarkers in cancer detection.. Medical Oncology 2016, 34, 7, 10.1007/s12032-016-0863-4.
