Cultural heritage objects constitute a very diverse environment, inhabited by various bacteria and fungi. The impact of these microorganisms on the degradation of artworks is undeniable, but at the same time, some of them may be applied for the efficient biotreatment of cultural heritage assets. Interventions with microorganisms have been proven to be useful in restoration of artworks, when classical chemical and mechanical methods fail or produce poor or short-term effects. The path to understanding the impact of microbes on historical objects relies mostly on multidisciplinary approaches, combining novel meta-omic technologies with classical cultivation experiments, and physico-chemical characterization of artworks.
- bacteria
- fungi
- biodeterioration
Note:All the information in this draft can be edited by authors. And the entry will be online only after authors edit and submit it.
1. Introduction
Cultural heritage objects represent highly heterologous habitats, both in terms of the microbiome’s structure and composition. Even though they are mostly considered as oligotrophic, these habitats can be colonized by various groups of microorganisms. Artworks made from textiles, paper, wood and even stones provide substrates for microbial growth, which often go unnoticed unless biofilm overgrowth and discoloration or weakening of the physical integrity of the material occurs. In consequence, many objects may lose some of their cultural and monetary value.
Much has been done to study the diversity of microorganisms responsible for biodeterioration, but there are still many unanswered questions. This lack of knowledge inhibits the development of effective and non-destructive conservation strategies. Since conventional physico-chemical methods are often ineffective and dangerous for treated artworks (as well as people involved in the conservation process), new strategies are constantly being developed to mitigate the negative impact of microorganisms. These include the use of gamma irradiation [1], vaporized hydrogen peroxide [2], low- temperature helium-generated plasm [3], volatile compounds and essential oils [4] as well as various natural biocides obtained from microorganisms, marine organisms or plants [5]. Many of them still need to be further verified, testing their usability in different study-case scenarios and with various materials. Nowadays, biocides are commonly used to limit biodeterioration of various objects. Amongst them, the application of nanoparticles is gaining increasing attention [5][6][5,6]. However, due to the highly penetrative nature of nanoparticles and their long-lasting effect on living organisms as well as their unknown interference with the treated material, their usage must be carefully monitored [7].
When historical artworks become damaged, intervention and conservation techniques need to be applied. A promising alternative for traditional strategies is the use of natural processes performed by microorganisms. Compounds of biological origin, such as calcium carbonate, can be used for biotreatment of damaged stones [8], while calcium oxalate films may act as an additional protective layer [9]. Biological treatment is characterized by low cost and low invasiveness, as well as high specificity and easy control when compared to traditional physico-chemical techniques [10]. Due to the multi- component nature of the decay, a combination of biological treatment and various other methods was also tested, with initial gentle treatment by mechanical, chemical or laser-based methods followed by the main cleaning process using microorganisms [11][12][13][11–13].
An important issue in cultural heritage microbiology is also public health concern. International human traffic within museums makes cultural heritage estates similar to some of the most crowded airports. In 2019, almost the same number of visitors (approximately 28 million) was recorded for the top five London museums combined (British Museum, Tate Modern, National Gallery, Natural History Museum, Victoria & Albert Museum) as for London Stansted Airport [14]. These make cultural heritage objects important bridge-nodes in the global network of pathogen spread. Therefore, the significance of such objects in pathogen transmission must not be underestimated and has to be further evaluated [15][16][17][15–17].
2. Inspection of Methodologies Applied for Identification and Characterization of Microorganisms in Cultural Heritage Objects
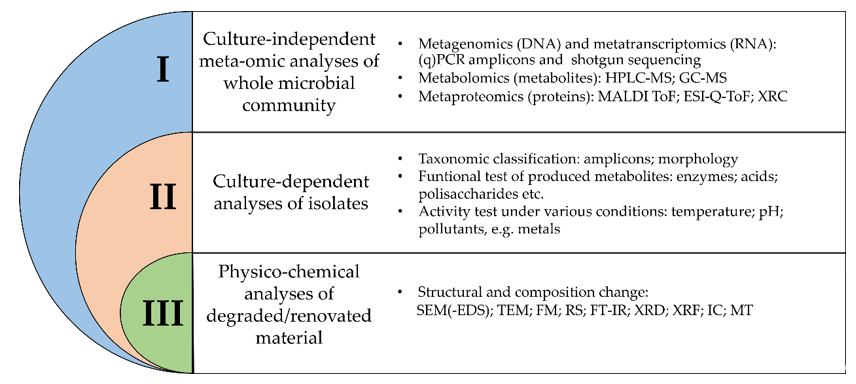
When studying objects of cultural heritage, three levels of complex microbiological research can be distinguished (Figure 1). Integration of all three approaches may provide a better understanding of microorganisms (including biodetriogens) inhabiting and influencing historical and cultural objects [18].
Figure 1. Overview of different methodologies for the study of microbial communities in relation to their impact on organic and inorganic materials. (q)PCR—(quantitative) polymerase chain reaction; HPLC-MS—liquid chromatography- mass spectrometry; GC-MS—gas chromatography-mass spectrometry; MALDI ToF—matrix-assisted laser desorption/ionization time-of-flight; ESI-Q-ToF—electrospray-ionization quadrupole time-of-flight mass spectrometry; XRC—X-ray crystallography; SEM(-EDS)—scanning electron microscopy (coupled with energy-dispersive X-ray spectroscopy); TEM—transmission electron microscopy; FM –fluorescence microscopy; RS—Raman spectroscopy; FT-IR—Fourier transform coupled infrared spectroscopy; XRD—X-ray diffraction; XRF—X-ray fluorescence; IC—ion chromatography; MT—microtomography.
Selection of an appropriate methodology for microbiological research is usually based on conservation restrictions and study purpose, such as general contamination (biodeterioration and human health risk assessment) or targeted research on a given artwork/material [19][20][21][22][23][19–23]. For general contamination assessment, the air is the most popular study subject, therefore numerous sampling techniques have been developed with (i) impaction, (ii) filtration, and (iii) impingement as major methods [24]. Microorganisms are collected onto an agar medium, filter and liquid medium, respectively. While impaction is a strictly culture-based approach, the other two may be culture-independent.
Although each method has its benefits, they also have their drawbacks. For example, the use of the culture-based approach, coupled with impaction, introduces a serious bias in the general biodiversity recovery rate. This is due to two main factors, i.e., the cultivation-based approach bias and the fact that if the agar medium is too solid, the cells can simply bounce back. The main drawback of the filtration method is low recovery efficiency and filter overloading [25]. The main disadvantage of impingementation is related to the evaporation of liquid. However, this may sometimes benefit the experiment, as it increases the concentration of the sample before the subsequent analyses [24][26][24,26].
When research on historic surfaces is conducted, only non-invasive or micro-invasive treatment methods can be applied using sterile scalpels, swabs, membrane filters or adhesive tapes [22][23][27][22,23,27]. However, it should be noted that the use of various sampling methods can provide different results. For example, nylon membranes are more reliable than cotton swabs, as the latter tend to overestimate the surface microbiota, which may not be involved in the biodeterioration process [27]. This stems from the fact that microbes tend to become endolytic in the search of nutrients but also need to protect themselves from UV radiation, temperature fluctuations and desiccation [28][29][30][31][28–31].
2.1. Culture-Independent Meta-Omic Analyses
Nowadays, metagenomic studies are often applied for the estimation of microbial contamination of objects of cultural heritage. Usually, the diversity of bacteria and fungi is analyzed by amplicon sequencing (metabarcoding), as this method requires a smaller amount of DNA and is cheaper than the whole metagenome sequencing.
The most common taxonomic markers are fragments of genes encoding ribosomal DNA (rDNA). In the case of bacteria, in particular, V3 and V4 hypervariable regions of the 16S rDNA gene are commonly used, as they provide the most reliable results [32][33][34][32–34]. In comparison, fungal diversity is usually studied using internal transcribed spacers (ITS), either ITS1 or ITS2. In general, the use of the ITS2 region is preferred as it was shown that the use of ITS1 can lead to overestimation of diversity and richness when compared to the ITS2 region [35]. However, a recent study showed that in the case of bio-aerosols the ITS1 region might be more suitable [36]. To avoid additional bias, such as chimeric and overrepresented sequences, the number of PCR cycles should be minimized and the use of proofreading DNA polymerase is highly recommended [37]. A maximum of thirty cycles of PCR should be sufficient for culture-dependent analyses, as well as culture- independent analyses of air and dust microbiomes.
There are many tools for the assessment of biodiversity based on amplicons. For example, the QIIME2 platform is extensively used for most bioinformatic analyses involved in the process of biodiversity research [38]. Various amplicon analysis tools have been already compared and reviewed elsewhere [39][40][39,40], but it is important to note that workflows based on Amplicon Sequence Variant (ASV), employed by DADA2, USEARCH-UNOISE3, and QIIME2, seem to be superior to the ones based on Operational Taxonomic Units.
Relatively easy, fast, reliable, and cost-effective quantitative assessment of microbial contamination could also be done by qPCR, targeting 16S rDNA gene, ITS regions or rbcl, tufA and actB genes [41][42][43][41–43]. Amplification primers, used both in PCR and qPCR reactions, usually cover specific groups of microorganisms, but degenerate primers, applicable to a broad range of taxa are used as well [42]. As mentioned earlier, in the case of bio-aerosols, a loss of material often occurs during sample processing, thus spiking and normalization of measurements needs to be done, allowing good correlations between qPCR and high- throughput sequencing [42].
A significant development in metagenomic studies came with the introduction of third-generation sequencing technologies. Most commonly used short-read based technologies (<300 bp) enable reliable biodiversity investigation on the genus level at the maximum, while third-generation sequencing technologies make it possible to analyze the data on species or even the strain level due to longer sequencing reads [44]. Recent researches show that many species-level assignments could be done only with long-read sequences, thus indicating the importance of improved resolution for accurate assessment of degradation/pathogenicity potential of environmental samples, usually observed for certain species or strains, but not always on the genera level [23][45][23,45].
Detailed characterization of microbial communities thriving on historical assets (in terms of both diversity and metabolic functions) can be done by whole metagenome sequencing [46][47][46,47]. However, this approach is rarely used in cultural heritage due to strict requirements of non-invasive or minimal-invasive sampling, which often prevents the required amount of DNA from being collected. Among available technologies, MinION devices (Oxford Nanopore system) exhibit high potential for application in the cultural heritage area as they require only a small amount of DNA. The small size of the device makes it suitable for on-site analyses. Moreover, longer reads gave a better insight into the relative proportions and metabolic potential of studied microbiomes [46]. The combination of various sequencing methods suitable for whole metagenome sequencing, such as the Oxford Nanopore, providing long reads but with lower accuracy when compared to the short but highly accurate reads provided by the Illumina MiSeq system, can significantly increase genome coverage (~200×) and therefore the quality of metagenome-derived genomes [48].
Although the majority of research related to cultural heritage microbiology is based on DNA sequencing, recent reports indicate that diversity analyses based on RNA molecules may provide better results as they focus on the identification of active microbes, rather than the overall microbial community [49]. Nevertheless, it should be noted that microorganisms often shift between the dormant and active state, depending on the environmental conditions, thus, when carrying out RNA-based analyses, sampling should be conducted multiple times, to prevent time-point bias of complex microbial communities [50][51][50,51]. RNA-based research is still rarely performed in cultural heritage microbiology, but there are already some interesting examples of total bacterial transcriptome analyses associated with historic artworks [52]. It is important to mention that one of the limitations of metatranscriptomic analyses is the lack of well-developed databases, but this may be overcome if metagenome-guided research is performed in parallel. In such a situation, metatranscriptomics can significantly contribute to the understanding of microbial processes occurring in the studied environment [53].
Among meta-omic approaches, other techniques have been recently introduced into the cultural heritage field. These novel methods are still rarely used and include metabolomic studies of wood and brick [54], wax seals [55], photographs [56], as well as metaproteomic studies of books [57], historical wood, frescoes and canvases [58][59][58,59], mummies and documents [60]. The metabolomic approach allows the investigation of many different compounds as it can detect and analyze proteins, lipids, and carbohydrates. Metabolomic studies may be either targeted, allowing for the analysis of a set of individual metabolites such as pigments, or untargeted (global), providing an insight into thousands of metabolites, which can be useful for screening for biomarkers of biological processes, such as biodeterioration.
2.2. Culture-Dependent Analyses
In cultural heritage assets, the most commonly used approach for monitoring of microorganisms is based on culture-dependent methods. Classical cultivation on agar media is simple, cheap, and can be easily applied by the basic-trained personnel belonging to museums. There are many different types of media with some specificity towards different taxonomic groups [61][62][63][61–63]. The main disadvantage of this approach is that the majority of microorganisms are not cultivable on classical media, which neglects the importance of viable but not culturable microorganisms [64]. Furthermore, while rich media are commonly used, they are not suitable for the overall biodiversity analyses [65]. Nevertheless, a cultivation approach can provide important data concerning microbial phenotypes, especially when prolonged incubation periods and low-substrate media are used for the increased recovery of environmental isolates [66][67][66,67]. Additionally, meta- omics aided cultivation strategies are promising for the isolation of yet uncultured microorganisms [68].
An undoubtful advantage of cultivable techniques is the enrichment of several microbial strains obtained in high cell density, which is crucial to perform laboratory assays (simulation assays or mitigation assays. Physiological analyses of single isolates may provide insight into the metabolic potential of a given microorganism (biodeteriogen), including its ability for pigment production, acid/alkaline production, proteolytic/cellulolytic activity or carbonate dissolution [69]. For example, in the work of Pavić and colleagues, it was shown that cultivable biodeteriogens were able to produce proteases, esterases, and lipases and grew on dyes as a sole source of phosphate and iron [70].
2.3. Physico-Chemical Analyses
Analyses of microorganisms associated with biodeterioration or biotreatment processes are more meaningful if auxiliary techniques, which visualize structural modifications or chemical changes, are applied. It has already been shown, that for complex multi-component analyses several commonly acknowledged techniques may be combined, e.g.,: (i) XRF, XRD, SEM, FM, isotope analyses, coupled with cultivation experiments on Reasoner’s 2A agar and 16S rDNA analyses (Applied Biosystems platform) [71]; (ii) SEM, RS, FTIR, together with cultivation experiments on Mueller– Hinton medium and 16S rDNA analyses (Applied Biosystems platform) [72]; (iii) XRD, RS, FTIR, SEM-EDS, confocal microscopy, accompanied with transcriptomic sequencing (Illumina) and shotgun sequencing (PacBio) [52].
Nowadays, some novel, non-invasive, analytical chemistry techniques are being applied in cultural heritage analyses. FTIR microspectroscopy and Raman spectroscopy are examples of non-destructive methods gaining increasing attention. These methods enable the monitoring of microbial contamination and the state of artwork by spectra identification. They have already been employed for the analyses of pigment [72], textile fibers [73][74][73,74], mineral formation [52], and other natural organic substances such as oils, gums or glues [75]. One of the newest methodologies, allowing for early detection of microorganism activity is a technique using RGB-ITR laser scanner (Red Green Blue Imaging Topological Radar) [76]. This approach is based on laser-induced fluorescence and 3D digitalization of the site and it can detect changes even in areas where biodeterioration is not yet evident.
The activity of microbes on artworks may be also assessed by ATP and FDA assays [77][78][79][77–79]. However, these activity assays should be applied only for rapid and simple testing as a preliminary evaluation before further analyses are performed [78].
References
- Drábková, K.; Ďurovič, M.; Kučerová, I. Influence of gamma radiation on properties of paper and textile fibres during disinfection. Phys. Chem. 2018, 152, 75–80, doi:10.1016/j.radphyschem.2018.07.023.
- Anna, W.; Dorota, R.; Mansur, R.; Sławomir, W. Microorganisms colonising historical cardboard objects from the Auschwitz-Birkenau State Museum in Oświęcim, Poland and their disinfection with vaporised hydrogen peroxide (VHP). Biodeterior. Biodegrad. 2020, 152, doi:10.1016/j.ibiod.2020.104997.
- Turnau, K.; Jędrzejczyk, R.J.; Ważny, R.; Chlebda, D.; Janicka, M.; Pawcenis, D.; Łojewski, T. Microbes of XVI century Arrases of Krakow Royal Castle. Res. 2020, 238, doi:10.1016/j.micres.2020.126485.
- Palla, F.; Bruno, M.; Mercurio, F.; Tantillo, A.; Rotolo, V. Essential oils as natural biocides in conservation of cultural heritage. Molecules 2020, 25, 730, doi:10.3390/molecules25030730.
- Fidanza, M.R.; Caneva, G. Natural biocides for the conservation of stone cultural heritage: A review. Cult. Herit. 2019, 38, 271–286, doi:10.1016/j.culher.2019.01.005.
- Kakakhel, M.A.; Wu, F.; Gu, J.D.; Feng, H.; Shah, K.; Wang, W. Controlling biodeterioration of cultural heritage objects with biocides: A review. Biodeterior. Biodegrad. 2019, 143, 104721, doi:10.1016/j.ibiod.2019.104721.
- Reyes-Estebanez, M.; Ortega-Morales, B.O.; Chan-Bacab, M.; Granados-Echegoyen, C.; Camacho-Chab, J.C.; Pereañez-Sacarias, J.E.; Gaylarde, C. Antimicrobial engineered nanoparticles in the built cultural heritage context and their ecotoxicological impact on animals and plants: a brief review. Sci. 2018, 6, 1–11, doi:10.1186/s40494-018-0219-9.
- Marvasi, M.; Mastromei, G.; Perito, B. Bacterial Calcium Carbonate Mineralization in situ Strategies for Conservation of Stone Artworks: From Cell Components to Microbial Community. Microbiol. 2020, 11, 1186.
- Rampazzi, L. Calcium oxalate films on works of art: A review. Cult. Herit. 2019, 40, 195–214.
- Soffritti, I.; D’Accolti, M.; Lanzoni, L.; Volta, A.; Bisi, M.; Mazzacane, S.; Caselli, E. The potential use of microorganisms as restorative agents: An update. Sustainability 2019, 11, 3853, doi:10.3390/su11143853.
- Troiano, F.; Gulotta, D.; Balloi, A.; Polo, A.; Toniolo, L.; Lombardi, E.; Daffonchio, D.; Sorlini, C.; Cappitelli, F. Successful combination of chemical and biological treatments forthecleaning of stone artworks. Biodeterior. Biodegrad. 2013, 85, 294–304, doi:10.1016/j.ibiod.2013.08.011.
- Rampazzo, M.; Manente, S.; Micheluz, A.; Ganzerla, R.; Ravagnan, G. How traces of pollutants in the environment modify bioremediation efficiency performed with Desulfovibrio vulgaris, and the advantage of an optimization protocol using soft chemicals. Cult. Herit. 2018, 30, doi:10.1016/j.culher.2017.09.017.
- Elhagrassy, A.F.; Hakeem, A.; Alhagrassy, A.F. Comparative Study of Biological Cleaning and Laser Techniques for Conservation of Weathered Stone in Failaka Island, Kuwait. Cult. 2018, 4, doi:10.5281/zenodo.1214561.
- Rubin, J.; Yin, T.; Au, G.; Chang, B.; Chee, M.; Cheu, L.; Datels, C.; Fischer, L.; Hoffman, M.; Kondaurova, O.; et al. TEA/AECOM 2018 Theme index and museum index: The global attractions attendance report. 2019, 1–90.
- Kim, K.H.; Kabir, E.; Jahan, S.A. Airborne bioaerosols and their impact on human health. Environ. Sci. 2018, 67, 23–35.
- Rascovan, N.; Telke, A.; Raoult, D.; Rolain, J.M.; Desnues, C. Exploring divergent antibiotic resistance genes in ancient metagenomes and discovery of a novel beta-lactamase family. Microbiol. Rep. 2016, 8, doi:10.1111/1758-2229.12453.
- Herfst, S.; Böhringer, M.; Karo, B.; Lawrence, P.; Lewis, N.S.; Mina, M.J.; Russell, C.J.; Steel, J.; de Swart, R.L.; Menge, C. Drivers of airborne human-to-human pathogen transmission. Opin. Virol. 2017, 22, 22–29.
- Adamiak, J.; Bonifay, V.; Otlewska, A.; Sunner, J.A.; Beech, I.B.; Stryszewska, T.; Kanka, S.; Oracz, J.; Zyzelewicz, D.; Gutarowska, B. Untargeted metabolomics approach in halophiles: Understanding the biodeterioration process of building materials. Microbiol. 2017, 8, doi:10.3389/fmicb.2017.02448.
- Dyda, M.; Pyzik, A.; Wilkojc, E.; Kwiatkowska-Kopka, B.; Sklodowska, A. Bacterial and fungal diversity inside the medieval building constructed with sandstone plates and lime mortar as an example of the microbial colonization of a nutrient-limited extreme environment (Wawel royal castle, Krakow, Poland). Microorganisms 2019, 7, 416, doi:10.3390/microorganisms7100416.
- Li, A.; Xiong, J.; Yao, L.; Gou, L.; Zhang, W. Determination of dust and microorganism accumulation in different designs of AHU system in Shaanxi History Museum. Environ. 2016, 104, doi:10.1016/j.buildenv.2016.05.014.
- Adamiak, J.; Otlewska, A.; Tafer, H.; Lopandic, K.; Gutarowska, B.; Sterflinger, K.; Piñar, G. First evaluation of the microbiome of built cultural heritage by using the Ion Torrent next generation sequencing platform. Biodeterior. Biodegrad. 2018, 131, 11–18, doi:10.1016/j.ibiod.2017.01.040.
- Liu, Z.; Zhang, Y.; Zhang, F.; Hu, C.; Liu, G.; Pan, J. Microbial community analyses of the deteriorated storeroom objects in the Tianjin Museum using culture-independent and culture-dependent approaches. Microbiol. 2018, 9, doi:10.3389/fmicb.2018.00802.
- Grottoli, A.; Beccaccioli, M.; Zoppis, E.; Fratini, R.S.; Schifano, E.; Santarelli, M.L.; Uccelletti, D.; Reverberi, M. Nanopore Sequencing and Bioinformatics for Rapidly Identifying Cultural Heritage Spoilage Microorganisms. Mater. 2020, 7, doi:10.3389/fmats.2020.00014.
- Ghosh, B.; Lal, H.; Srivastava, A. Review of bioaerosols in indoor environment with special reference to sampling, analysis and control mechanisms. Int. 2015, 85, 254–272.
- Bøifot, K.O.; Gohli, J.; Moen, L.V.; Dybwad, M. Performance evaluation of a new custom, multi-component DNA isolation method optimized for use in shotgun metagenomic sequencing-based aerosol microbiome research. Microbiomes 2020, 15, doi:10.1186/s40793-019-0349-z.
- Ferguson, R.M.W.; Garcia-Alcega, S.; Coulon, F.; Dumbrell, A.J.; Whitby, C.; Colbeck, I. Bioaerosol biomonitoring: Sampling optimization for molecular microbial ecology. Ecol. Resour. 2019, 19, doi:10.1111/1755-0998.13002.
- Piñar, G.; Tafer, H.; Sterflinger, K.; Pinzari, F. Amid the possible causes of a very famous foxing: Molecular and microscopic insight into Leonardo da Vinci’s self-portrait. Microbiol. Rep. 2015, 7, doi:10.1111/1758-2229.12313.
- Goyal, P.; Bhola, D.; Varma, A. Introduction to modern tools and techniques to understand microbes. In Modern Tools and Techniques to Understand Microbes; Springer, Cham, Switzerland, 1–23.
- Gutarowska, B.; Pietrzak, K.; Machnowski, W.; Milczarek, J.M. Historical textiles – a review of microbial deterioration analysis and disinfection methods. Res. J. 2017, 87, 2388–2406.
- Sanmartín, P.; DeAraujo, A.; Vasanthakumar, A. Melding the Old with the New: Trends in Methods Used to Identify, Monitor, and Control Microorganisms on Cultural Heritage Materials. Ecol. 2018, 76, 64–80, doi:10.1007/s00248-016-0770-4.
- Gutarowska, B. The use of -omics tools for assessing biodeterioration of cultural heritage: A review. Cult. Herit. 2020, 351–361 doi:10.1016/j.culher.2020.03.006
- Adams, R.I.; Bateman, A.C.; Bik, H.M.; Meadow, J.F. Microbiota of the indoor environment: A meta-analysis. Microbiome 2015, 3, doi:10.1186/s40168-015-0108-3.
- Barb, J.J.; Oler, A.J.; Kim, H.S.; Chalmers, N.; Wallen, G.R.; Cashion, A.; Munson, P.J.; Ames, N.J. Development of an Analysis Pipeline Characterizing Multiple Hypervariable Regions of 16S rRNA Using Mock Samples. PLoS ONE 2016, 11, doi:10.1371/journal.pone.0148047.
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P.D. 16S rRNA gene sequencing of mock microbial populations-impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol. 2016, 16, doi:10.1186/s12866-016-0738-z.
- Yang, R.H.; Su, J.H.; Shang, J.J.; Wu, Y.Y.; Li, Y.; Bao, D.P.; Yao, Y.J. Evaluation of the ribosomal DNA internal transcribed spacer (ITS), specifically ITS1 and ITS2, for the analysis of fungal diversity by deep sequencing. PLoS ONE 2018, 13, doi:10.1371/journal.pone.0206428.
- Mbareche, H.; Veillette, M.; Bilodeau, G.; Duchaine, C. Comparison of the performance of ITS1 and ITS2 as barcodes in amplicon-based sequencing of bioaerosols. PeerJ 2020, 2020, doi:10.7717/peerj.8523.
- Sze, M.A.; Schloss, P.D. The Impact of DNA Polymerase and Number of Rounds of Amplification in PCR on 16S rRNA Gene Sequence Data. mSphere 2019, 4, doi:10.1128/msphere.00163-19.
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Biotechnol. 2019, 37, 852–857, doi:10.1038/s41587-019-0209-9.
- Pauvert, C.; Buée, M.; Laval, V.; Edel-Hermann, V.; Fauchery, L.; Gautier, A.; Lesur, I.; Vallance, J.; Vacher, C. Bioinformatics matters: The accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline. Fungal Ecol. 2019, 41, doi:10.1016/j.funeco.2019.03.005.
- Prodan, A.; Tremaroli, V.; Brolin, H.; Zwinderman, A.H.; Nieuwdorp, M.; Levin, E. Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS ONE 2020, 15, doi:10.1371/journal.pone.0227434.
- Ettenauer, J.; Piñar, G.; Tafer, H.; Sterflinger, K. Quantification of fungal abundance on cultural heritage using real time PCR targeting the β-actin gene. Microbiol. 2014, 5, doi:10.3389/fmicb.2014.00262.
- Unterwurzacher, V.; Pogner, C.; Berger, H.; Strauss, J.; Strauss-Goller, S.; Gorfer, M. Validation of a quantitative PCR based detection system for indoor mold exposure assessment in bioaerosols. Sci. Process. Impacts 2018, 20, 1454–1468, doi:10.1039/c8em00253c.
- del Mondo, A.; de Natale, A.; Pinto, G.; Pollio, A. Correction to: Novel qPCR probe systems for the characterization of subaerial biofilms on stone monuments. Microbiol. 2019, 69, doi:10.1007/s13213-019-01480-9.
- Šoltys, K.; Planý, M.; Biocca, P.; Vianello, V.; Bučková, M.; Puškárová, A.; Sclocchi, M.C.; Colaizzi, P.; Bicchieri, M.; Pangallo, D.; et al. Lead soaps formation and biodiversity in a XVIII Century wax seal coloured with minium. Microbiol. 2020, 22, doi:10.1111/1462-2920.14735.
- Nygaard, A.B.; Tunsjø, H.S.; Meisal, R.; Charnock, C. A preliminary study on the potential of Nanopore MinION and Illumina MiSeq 16S rRNA gene sequencing to characterize building-dust microbiomes. Rep. 2020, 10, doi:10.1038/s41598-020-59771-0.
- Piñar, G.; Poyntner, C.; Lopandic, K.; Tafer, H.; Sterflinger, K. Rapid diagnosis of biological colonization in cultural artefacts using the MinION nanopore sequencing technology. Biodeterior. Biodegrad. 2020, 148, doi:10.1016/j.ibiod.2020.104908.
- Louati, M.; Ennis, N.J.; Ghodhbane-Gtari, F.; Hezbri, K.; Sevigny, J.L.; Fahnestock, M.F.; Cherif-Silini, H.; Bryce, J.G.; Tisa, L.S.; Gtari, M. Elucidating the ecological networks in stone-dwelling microbiomes. Microbiol. 2020, 22, doi:10.1111/1462-2920.14700.
- Bertrand, D.; Shaw, J.; Kalathiyappan, M.; Ng, A.H.Q.; Kumar, M.S.; Li, C.; Dvornicic, M.; Soldo, J.P.; Koh, J.Y.; Tong, C.; et al. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Biotechnol. 2019, 37, doi:10.1038/s41587-019-0191-2.
- Meng, H.; Zhang, X.; Katayama, Y.; Ge, Q.; Gu, J.D. Microbial diversity and composition of the Preah Vihear temple in Cambodia by high-throughput sequencing based on genomic DNA and RNA. Biodeterior. Biodegrad. 2020, 149, doi:10.1016/j.ibiod.2020.104936.
- Van Vliet, S. Bacterial Dormancy: How to Decide When to Wake Up. Biol. 2015, 25, R753-5.
- Villa, F.; Vasanthakumar, A.; Mitchell, R.; Cappitelli, F. RNA-based molecular survey of biodiversity of limestone tombstone microbiota in response to atmospheric sulphur pollution. Appl. Microbiol. 2015, 60, 92–102, doi:10.1111/lam.12345.
- Kooli, W.M.; Junier, T.; Shakya, M.; Monachon, M.; Davenport, K.W.; Vaideeswaran, K.; Vernudachi, A.; Marozau, I.; Monrouzeau, T.; Gleasner, C.D.; et al. Remedial treatment of corroded iron objects by environmental Aeromonas isolates. Environ. Microbiol. 2019, 85, doi:10.1128/AEM.02042-18.
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and challenges in metatranscriptomic analysis. Genet. 2019, 10, 904.
- Gutarowska, B.; Celikkol-Aydin, S.; Bonifay, V.; Otlewska, A.; Aydin, E.; Oldham, A.L.; Brauer, J.I.; Duncan, K.E.; Adamiak, J.; Sunner, J.A.; et al. Metabolomic and high-throughput sequencing analysis-modern approach for the assessment of biodeterioration of materials from historic buildings. Microbiol. 2015, 6, doi:10.3389/fmicb.2015.00979.
- Szulc, J.; Jablonskaja, I.; Jabłońska, E.; Ruman, T.; Karbowska-Berent, J.; Gutarowska, B. Metabolomics and metagenomics characteristic of historic beeswax seals. Biodeterior. Biodegrad. 2020, 152, doi:10.1016/j.ibiod.2020.105012.
- Szulc, J.; Ruman, T.; Karbowska-Berent, J.; Kozielec, T.; Gutarowska, B. Analyses of microorganisms and metabolites diversity on historic photographs using innovative methods. Cult. Herit. 2020, doi:10.1016/j.culher.2020.04.017.
- Kraková, L.; Šoltys, K.; Otlewska, A.; Pietrzak, K.; Purkrtová, S.; Savická, D.; Puškárová, A.; Bučková, M.; Szemes, T.; Budiš, J.; et al. Comparison of methods for identification of microbial communities in book collections: Culture-dependent (sequencing and MALDI-TOF MS) and culture-independent (Illumina MiSeq). Biodeterior. Biodegrad. 2018, 131, doi:10.1016/j.ibiod.2017.02.015.
- Manfredi, M.; Barberis, E.; Gosetti, F.; Conte, E.; Gatti, G.; Mattu, C.; Robotti, E.; Zilberstein, G.; Koman, I.; Zilberstein, S.; et al. Method for Noninvasive Analysis of Proteins and Small Molecules from Ancient Objects. Chem. 2017, 89, doi:10.1021/acs.analchem.6b03722.
- Barberis, E.; Baiocco, S.; Conte, E.; Gosetti, F.; Rava, A.; Zilberstein, G.; Righetti, P.G.; Marengo, E.; Manfredi, M. Towards the non-invasive proteomic analysis of cultural heritage objects. J. 2018, 139, doi:10.1016/j.microc.2018.03.033.
- Giuffrida, M.G.; Mazzoli, R.; Pessione, E. Back to the past: deciphering cultural heritage secrets by protein identification. Microbiol. Biotechnol. 2018, 102, 5445–5455.
- Atlas, R. Handbook of Microbiological Media, 4th ed.; CRC press: Boca Raton, FL, USA, 2010.
- Basu, S.; Bose, C.; Ojha, N.; Das, N.; Das, J.; Pal, M.; Khurana, S. Evolution of Bacterial and Fungal Growth Media. Bioinformation, 2015, 11, 182–184, doi: 10.6026/97320630011182
- Bonnet, M.; Lagier, J.C.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: the evolution of culture media in clinical microbiology. New Microbes New Infect. 2020, 34, 100622.
- Laiz, L.; Piñar, G.; Lubitz, W.; Saiz-Jimenez, C. Monitoring the colonization of monuments by bacteria: Cultivation versus molecular methods. Microbiol. 2003, 5, doi:10.1046/j.1462-2920.2003.00381.x.
- Dziurzynski, M.; Ciuchcinski, K.; Dyda, M.; Szych, A.; Drabik, P.; Laudy, A.; Dziewit, L. Assessment of bacterial contamination of air at the museum of King John III’s palace at wilanow (Warsaw, Poland): Selection of an optimal growth medium for analyzing airborne bacteria diversity. Sci. 2020, 10, 7128, doi:10.3390/app10207128.
- Pangallo, D.; Bučková, M.; Kraková, L.; Puškárová, A.; Šaková, N.; Grivalský, T.; Chovanová, K.; Zemánková, M. Biodeterioration of epoxy resin: A microbial survey through culture-independent and culture-dependent approaches. Microbiol. 2015, 17, doi:10.1111/1462-2920.12523.
- Chaudhary, D.K.; Khulan, A.; Kim, J. Development of a novel cultivation technique for uncultured soil bacteria. Rep. 2019, 9, doi:10.1038/s41598-019-43182-x.
- Gutleben, J.; Chaib De Mares, M.; van Elsas, J.D.; Smidt, H.; Overmann, J.; Sipkema, D. The multi-omics promise in context: from sequence to microbial isolate. Rev. Microbiol. 2018, 44, 212–229.
- Savković, Ž.; Stupar, M.; Unković, N.; Ivanović, Ž.; Blagojević, J.; Vukojević, J.; Ljaljević Grbić, M. In vitro biodegradation potential of airborne Aspergilli and Penicillia. Nat. 2019, 106, doi:10.1007/s00114-019-1603-3.
- Pavić, A.; Ilić-Tomić, T.; Pačevski, A.; Nedeljković, T.; Vasiljević, B.; Morić, I. Diversity and biodeteriorative potential of bacterial isolates from deteriorated modern combined-technique canvas painting. Biodeterior. Biodegrad. 2015, 97, doi:10.1016/j.ibiod.2014.11.012.
- Lepinay, C.; Mihajlovski, A.; Touron, S.; Seyer, D.; Bousta, F.; Di Martino, P. Bacterial diversity associated with saline efflorescences damaging the walls of a French decorated prehistoric cave registered as a World Cultural Heritage Site. Biodeterior. Biodegrad. 2018, 130, doi:10.1016/j.ibiod.2018.03.016.
- Cojoc, L.R.; Enache, M.I.; Neagu, S.E.; Lungulescu, M.; Setnescu, R.; Ruginescu, R.; Gomoiu, I. Carotenoids produced by halophilic bacterial strains on mural paintings and laboratory conditions. FEMS Microbiol. Lett. 2019, 366, doi:10.1093/femsle/fnz243.
- Kavkler, K.; Gunde Cimerman, N.; Zalar, P.; Demšar, A. FT-Raman analysis of cellulose based museum textiles: Comparison of objects infected and non-infected by fungi. Tekstilec 2018, 61, doi:10.14502/TEKSTILEC2018.61.110-123.
- Margariti, C. The application of FTIR microspectroscopy in a non-invasive and non-destructive way to the study and conservation of mineralised excavated textiles. Sci. 2019, 7, doi:10.1186/s40494-019-0304-8.
- Daher, C.; Bellot-Gurlet, L.; Le Hô, A.S.; Paris, C.; Regert, M. Advanced discriminating criteria for natural organic substances of Cultural Heritage interest: Spectral decomposition and multivariate analyses of FT-Raman and FT-IR signatures. Talanta 2013, 115, doi:10.1016/j.talanta.2013.06.014.
- Caneve, L.; Guarneri, M.; Lai, A.; Spizzichino, V.; Ceccarelli, S.; Mazzei, B. Non-destructive laser based techniques for biodegradation analysis in cultural heritage. NDT E Int. 2019, 104, doi:10.1016/j.ndteint.2019.03.007.
- Troiano, F.; Polo, A.; Villa, F.; Cappitelli, F. Assessing the microbiological risk to stored sixteenth century parchment manuscripts: A holistic approach based on molecular and environmental studies. Biofouling 2014, 30, doi:10.1080/08927014.2013.871539.
- Unković, N.; Ljaljević Grbić, M.; Stupar, M.; Vukojević, J.; Subakov-Simić, G.; Jelikić, A.; Stanojević, D. ATP bioluminescence method: tool for rapid screening of organic and microbial contaminants on deteriorated mural paintings. Prod. Res. 2019, 33, doi:10.1080/14786419.2015.1108975.
- Sirt Çıplak, E.; Kiraz, G.A. Enzymatic Activity as a Measure of Total Microbial Activity on Historical Stone. Heritage 2020, 3, 671–681, doi:10.3390/heritage3030038.