The ligand of Numb protein-X (LNX) family, also known as the PDZRN family, is composed of four discrete RING-type E3 ubiquitin ligases (LNX1, LNX2, LNX3, and LNX4), and LNX5 which may not act as an E3 ubiquitin ligase owing to the lack of the RING domain.
- LNX
- PDZRN
- RING
- PDZ domain,E3 ubiquitin ligase,UPS,lysosomal pathway,disease
- PDZRN
- RING
- PDZ domain,E3 ubiquitin ligase,UPS,lysosomal pathway,disease
Dear author, the following contents are excerpts from your papers. They are editable. And the entry will be online only after authors edit and submit it.
1. Introduction
Protein ubiquitylation, which is a highly conserved post-translational modification in which ubiquitin (consisting of 76 amino acids) is covalently linked to lysine residues of a substrate protein, is associated with nearly all aspects of eukaryotic physiology. The number of genes encoding proteins involved in ubiquitin-dependent post-translational modification in humans is estimated to be over 1000, which far exceed the total number of kinase-encoding genes (518) identified thus far [1,2][1][2]. Protein ubiquitylation comprises a series of enzymatic reactions that initially uses energy from ATP hydrolysis and is mediated by ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3), which ultimately render substrate specificity [3,4][3][4]. E3 ubiquitin ligases can be classified into three groups based on the presence of discrete domains. While HECT family of E3 ubiquitin ligases acts as a genuine enzyme by exploiting its catalytic cysteine residue located in C-terminal HECT domain, the E3 ligase with RING or U-box domain serves as a scaffold that recruits the E2 enzyme in close proximity to the bound substrate. While protein turnover mediated by the UPS is essential for cellular proteostasis, nonproteolytic ubiquitin modification is important for the regulation of epigenetic control, transcriptional regulation, cellular trafficking, and post-translational modification [5][5]. The presence of extracellular 20S proteasome in blood plasma and cancer cell culture conditioned media indicates the complexity of proteostasis [6][6]. Therefore, it is not surprising that UPS dysfunction is associated with multiple human diseases. Thus far enormous efforts have been dedicated to understanding the pathophysiological roles of individual E3s because the complexity of the UPS mostly stems from multiple substrates or diverse cellular roles of a single substrate of an E3 ubiquitin ligase [7,8][7][8].
Here we describe one of the multifunctional E3 ubiquitin ligases family, ligand of numb protein-X (LNX/PDZRN). Among the five members of the LNX family (LNX1-LNX5), LNX1-LNX4 may function as E3 ubiquitin ligases that target miscellaneous substrates for ubiquitylation, as these possess a RING domain with multiple PDZ (PSD95, DLGA, ZO-1) domains, each of which are considered protein-protein interacting modules [9,10,11,12,13][9][10][11][12][13]. Although over 600 E3 ubiquitin ligases are encoded in the human genome [1][1], LNX proteins belong to the sole E3 ligase family with multiple PDZ domains (up to four); therefore, it is not surprising that members of the LNX family appear to interact promiscuously with various different proteins [14,15,16,17][14][15][16][17]. Among the LNX proteins, two were originally named LNX1 (also called PDZRN2) and LNX2 (PDZRN1) owing to their ability to bind to NUMB, an intracellular Notch inhibitor[18] [18] via relatively well-conserved NUMB-binding motifs (NPAY in LNX1, NPAF in LNX2) located between the RING domain and the first PDZ domain; the consensus sequences from these are not present in other members of the LNX family. Thus LNX3, LNX4, and LNX5 are more prone to be referred to as PDZRN3, PDZRN4, and PDZRN5 (or PDZRN4L), respectively though (PDZRN; PDZ and RING) [9,17,19,20,21][9][17][19][20][21], we will collectively call the several synonyms of LNX/PDZRN/SEMCAP as LNX hereafter.
Although considerable efforts have been devoted during the last two decades to elucidating the function of LNXs, our knowledge of their substrates, binding partners, pathophysiological functions, and detailed molecular mechanisms of action of LNX proteins remains elusive. As an indication of the increased interest in this area of research, an excellent review article published in 2018 [17] [17] comprehensively discussed the role of LNX1 and LNX2 in various diseases. Therefore, in this review, after primarily emphasizing novel discoveries on LNX1 and LNX2 in the last three years, we will focus on findings of the cellular and pathogenic roles of LNX3 and LNX4, and then discuss what major questions remain to be tackled.
2. Structural Significance and Sequence Similarity of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family
Although the complete X-ray crystallographic structure of LNX proteins is not available, fragmented structures of LNX1p80 and LNX2 are published on various web sites and in the literature. According to previous reports based on the available X-ray crystallographic structures, the N-terminal three-dimensional structures of LNX1 and LNX2 show the presence of a single RING domain with two Zn finger motifs flanking the ends of the RING domain (Zn-RING-Zn fragment), and LNX2 s PDZ domain [22,23,24][22][23][24]. Structural information on the other parts of individual PDZ domains of LNX1 (PDZ2 and PDZ3) are also available in the Protein Data Bank (PDB; http://www.rcsb.org). In 2018 Young [17] [17]provided a detailed account of the structural information of LNX1 and LNX2 that is available. The predicted amino acid sequences and primary structures of human LNX proteins, with domain annotations, are shown in Figure 1. RING-type E3 ubiquitin ligases tend to form homodimers or heterodimers to acquire enzymatic properties, though some act as monomers [25][25]. Although both LNX1 and LNX2 Zn-RING-Zn fragments could be homodimerized, homodimerization is exclusively required for LNX1 to function as an E3 ubiquitin ligase, while the monomeric status of LNX2 is sufficient for the retention of its auto-ubiquitylation activity [22,24][22][24].
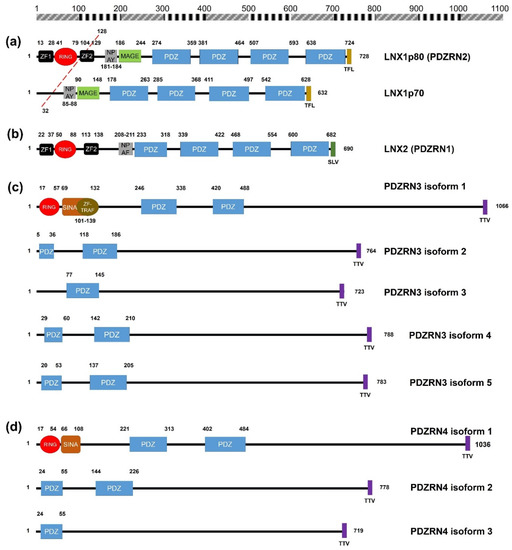
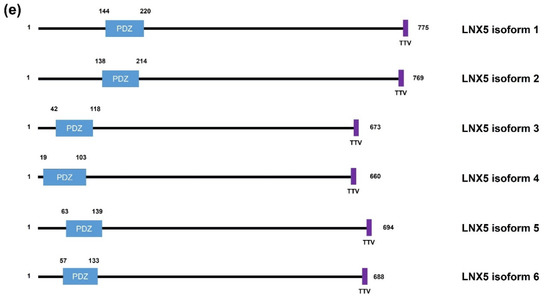
Figure 1. Schematic diagrams of the members of the human LNX family. (a) LNX1 has two splicing variants, LNX1p70 and LNX1p80. In contrast to the longer splicing variant, the shorter one does not contain the N-terminal RING domain and the two neighboring Zn finger motifs, while the other known domains following the second Zn finger motif are conserved, which include four PDZ domains and a NUMB-binding motif. Both variants bind to NUMB via the NPAY motif and contain a MAGE-B interacting region. The red dashed bar indicates the point from which the amino acid sequences become identical. (b) LNX2 showed the highest similarity with LNX1p80 in terms of primary structure. However, the major difference between LNX1p80 and LNX2 was noted at the region located between the NUMB-binding motif and the point of initiation of the first PDZ domain. While LNX1p80 has a relatively longer sequence that includes the MAGE-B18-interacting motif, LNX2 only has a 21-amino acid-spanning region. (c) LNX3 has five splicing variants. The PDZRN4 isoform 1 is the sole variant equipped with an N-terminal RING domain and a juxtaposed Zn finger flanking the C-terminal end of the RING domain. All the variants have two PDZ domains, except the third isoform, which only has one PDZ domain. (d) Among LNX4 variants, which include three isoforms, only the PDZRN4 isoform 1 has an N-terminal RING domain and a SINA-like motif. Similar to LNX3, the variants have two PDZ domains; the third isoform is an exception as it has only one relatively short PDZ domain identical to that in the PDZRN4 isoform 3. (e) LNX5 has six splicing variants. All variants have a single PDZ without any recognizable domains. The last three single letter amino acid codes represent the class I PDZ-binding motif (S/T-X-V/I/L). The scale bar on the top indicates the approximate length in terms of the number of amino acids for the comparison of individual proteins. ZF; Zn finger motif, RING; really interesting new gene, MAGE; melanoma-associated antigen, PDZ; PSD95-DLGA-ZO-1, SINA; seven in absentia, TRAF; tumor necrosis factor receptor (TNF-R)-associated factor. The schematic illustration of the alignment of the LNX/PDZRN domains are derived from the deduced amino acid sequence information retrieved from GenBank; the corresponding accession numbers have been provided: LNX1p80 (NP_001119800.1), LNX1p70 (NP_116011.2), LNX2 (NP_699202.1), PDZRN3 isoform 1 (NP_055824.1), PDZRN3 isoform 2 (NP_001290068.1), PDZRN3 isoform 3 (NP_001290069.1), PDZRN3 isoform 4 (NP_001290070.1), PDZRN3 isoform 5 (NP_001290071.1), PDZRN4 isoform 1 (NP_001158067.1), PDZRN4 isoform 2 (NP_037509.3), PDZRN4 isoform 3 (EAW57827.1), LNX5 isoform 1 (NP_001290441.1), LNX5 isoform 2 (NP_115901.2), LNX5 isoform 3 (NP_001290442.1), LNX5 isoform 4 (NP_001290443.1), LNX5 isoform 5 (NP_001290444.1), LNX5 isoform 6 (NP_001290445.1).
The possible formation of an oligomer or heteromer between LNX1 and LNX2 may be considered if we take into account the existence of multiple PDZ domains and class I PDZ-binding motifs (S/T-X-V/I/L) at the C-termini of both proteins (Figure 1a,b). The oligo- or heterodimeric complex formation was validated in yeast two-hybrid experiments, where the presence of C-terminal class I PDZ-binding motifs were a prerequisite for studying their intermolecular interaction [9][9]. However, whether the Zn-RING-Zn domains of both LNX1 and LNX2 can mediate heterodimerization has not been investigated thus far. Collectively, the dimerization sites in LNX1 and LNX2 may exist in at least two different positions: in the N-terminus containing the RING and Zn finger motifs, and in the PDZ domains. The proposed concepts were strongly supported by the finding that while the introduction of multiple mutations in the gene encoding the isolated Zn-RING-Zn domain was adequate to disrupt homodimerization of the fragments, when introduced in the genes encoding full-length LNX1 and LNX2 the same mutations failed to prevent dimerization [22,24][22][24]. Additionally, the intramolecular loop of the C-terminal class I PDZ-binding motifs could also have bound to the second PDZ domains (class I PDZ domain is able to bind S/T-X-C) of LNX1 and LNX2 [15][15]. Therefore, their intrinsic E3 ubiquitin ligase functions were regulated at the intramolecular level, and could be further modulated by post-translational modifications, such as by the attachment of phosphate moieties. Thus far, only a single protein kinase (c-Src) has been reported to mediate LNX1p80 phosphorylation [26][26]. In addition, since LNX family besides LNX5 has several Zn ion coordination sites, it would be encouraged to establish the optimal level of Zn ion to achieve as fully active recombinant proteins as possible in vitro by following well established experimental procedures [27,28][27][28].
Besides LNX5, the remaining members of the LNX family (PDZRN3 and PDZRN4) contain an N-terminal RING domain and two tandem repeat PDZ domains (Figure 1c–e). While PDZRN3 contains a Zn finger motif at the C-terminal end of its RING domain [29][29], the presence of this motif in PDZRN4 could not be confirmed, although PDZRN4 does contain a SINA-like domain that may incorporate a Zn ion. Interestingly, the sequences of all members of the LNX family (except LNX1 and LNX2), including their variants, end with a TTV segment, which is also a type of class I PDZ-binding motif. Therefore, LNX1 and LNX2 might interact with other LNX proteins via their PDZ domains and C-terminal class I PDZ-binding motifs. Alternative assumptions might be also possible, considering the structural homology could be inferred from the level of amino acid sequence similarity. Strikingly by pairwise comparison of the individual members of LNX/PDZRN family (Figure 2), we only observed the high sequence similarities between LNX1 and LNX2, and LNX3 and LNX4. While LNX1 and LNX2 as their names imply share some substrates (Table 2) including Numb for ubiquitin dependent modification, LNX3 (PDZRN3) and LNX4 (PDZRN4) though their overlapping targets has not been reported yet would possibly be complementary to each other at least in part under the particular cellular contexts by sharing common substrates. To clarify the unique or overlapping molecular and cellular functions of the individual LNX proteins, their mutually complementary roles on possible target proteins must be scrutinized through future studies.
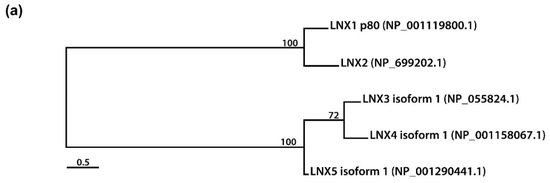
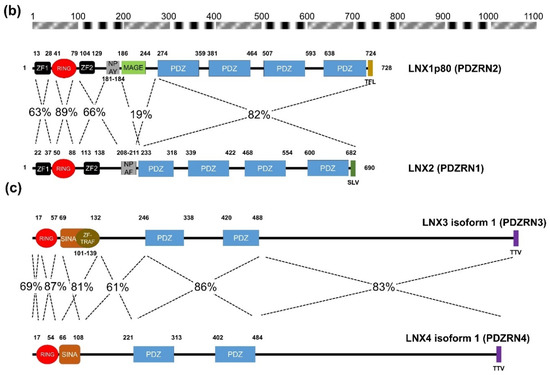
Figure 2. Amino acid sequence comparison of human LNXs (a) Phylogenetic tree of the LNX proteins. Maximum likelihood analysis was performed using to RAxML under the LG+GAMMA model. Analysis was carried out with five LNXs encoding the longest of their own variants (LNX1p80, LNX2, LNX3 isoform 1, LNX4 isoform 1, and LNX5 isoform 1). The numbers on each node represent bootstrap supported values. The scale bar indicates the number of substitutions/site. (b,c) Homology comparison of each domains was highlighted by the value of percentage. (b) Pairwise comparison between LNX1 and LNX2. The scale bar on the top indicates the approximate numbers of amino acids for the comparison of individual proteins. (c) Pairwise comparison between LNX3 (PDZRN3) and LNX4 (PDZRN4).
Table 2. Newly discovered LNX1 and LNX2 associated proteins in the past three years (2018–2020).
| Associated Proteins | LNX | Domain Involved | Methods Used | Major Functions | Consequences | References |
|---|
| NEK6 |
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle’s Dynamics and Signaling. PLoS ONE 2008, 3, e1487.
| LNX1 | ||||||
| RING and 3rd PDZ | Co-IP, GST pull down, Yeast two hybrid, | Serine/threonine kinase | K48-linked polyubiquitylated NEK6 undergoes UPS-dependent degradation | [31] (2020) | ||
| LDOC 1 | LNX1 | Co-IP | Tumor suppressor | LNX1 uses LDOC1 as a scaffold protein to indirectly target phospho-JAK2 destruction | [32] (2019) | |
| MDM2 | LNX1 | Co-IP | E3 ubiquitin ligase | LNX1 may indirectly interact with MDM2 | [ | |
| MDM2 | ||||||
| 37 | ] (2019) | |||||
| LNX1 | Co-IP | E3 ubiquitin ligase | LNX1 may indirectly interact with MDM2 | [37](2019) | ||
| p53 | LNX1 | Co-IP | Tumor suppressor | LNX1 indirectly mediates p53 destruction | [37] (2019) | |
| p53 | LNX1 | Co-IP | Tumor suppressor | LNX1 indirectly mediates p53 destruction | [37](2019) | |
| Connexin 36 | LNX1 | 2nd PDZ | Co-IP, GST pull down, Ni-NTA pull down | Gap junction protein | Ubiquitylated connexin36 undergoes lysosomal-dependent degradation. | [50] (2018) |
| Connexin 36 | LNX2 | 2nd PDZ | Co-IP, Ni-NTA pull down | Gap junction protein | Ubiquitylated connexin36 undergoes lysosomal-dependent degradation. | [50] (2018) |
| Connexin 36 | LNX2 | 2nd PDZ | Co-IP, Ni-NTA pull down | Gap junction protein | Ubiquitylated connexin36 undergoes lysosomal-dependent degradation. | [50](2018) |
| EphB1 | LNX1 p70 | N-terminal region ahead of 1st PDZ domain | Co-IP | Receptor tyrosine kinase | Stabilization | [56] (2018) |
| EphB1 | LNX1 p70 | N-terminal region ahead of 1st PDZ domain | Co-IP | Receptor tyrosine kinase | Stabilization | [56](2018) |
| EphB2 | LNX1 p80 | 56] (2018) | ||||
| EphB2 | LNX1 p80 | |||||
| Receptor tyrosine kinase | Degradation | |||||
| Receptor tyrosine kinase | ||||||
| [ | ||||||
| Degradation | [ | 56 | ] | (2018) | ||
| EphB2 | LNX1 p70 | 2nd PDZ | Co-IP, GST-pull down | Receptor tyrosine kinase | 57] (2019) | |
| EphB2 | LNX1 p70 | |||||
| Stabilization | ||||||
| 2nd PDZ | ||||||
| [ | 56 | ] (2018), | ||||
| Co-IP, GST-pull down | ||||||
| Receptor tyrosine kinase | ||||||
| [ | ||||||
| Stabilization | [ | 56 | ] | (2018), [57] (2019) |
||
| GluN1 | LNX1 | [ | 57] (2019) | |||
| GluN1 | LNX1 | |||||
| Co-IP | Glutamate receptor subunit | |||||
| Co-IP | Glutamate receptor subunit | LNX1 helps form a NMDAR complex by recruiting GluN1 and GluN2B | [57](2019) | |||
| GluN2B | LNX1 | 1st PDZ | Co-IP, GST pull down | Glutamate receptor subunit | LNX1 helps form a NMDAR complex by recruiting GluN1 and GluN2B | [ |
| GluN2B | ||||||
| 57 | ] (2019) | |||||
| LNX1 | 1st PDZ | Co-IP, GST pull down | Glutamate receptor subunit | LNX1 helps form a NMDAR complex by recruiting GluN1 and GluN2B | [57](2019) | |
| GlyT2 | LNX1, LNX2 | 2nd PDZ | ||||
| 2nd PDZ |
- Clague, M.J.; Heride, C.; Urbé, S. The demographics of the ubiquitin system. Trends Cell Biol.
| Co-IP | ||
| Glycine transporter | ||
| Polyubiquitylated and degraded | ||
| [ | ||
| 58 | ||
| ] (2019) | ||
| GlyT2 | LNX1, LNX2 | Co-IP |
- 2015
| Glycine transporter |
| Polyubiquitylated and degraded |
| [ |
| 58 |
| ] |
| (2019) |
- ,
- 25
| LNX1 helps form a NMDAR complex by recruiting GluN1 and GluN2B |
- , 417–426.
- Lipkowitz, S.; Weissman, A.M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 2011, 11, 629–643.
- Ro, H.; Hur, T.-L.; Rhee, M. Ubiquitin conjugation system for body axes specification in vertebrates. Anim. Cells Syst. 2015, 19, 87–95.
- Bhat, K.P.; Greer, S.F. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2011, 1809, 150–155.
- Tsimokha, A.S.; Artamonova, T.O.; Diakonov, E.E.; Khodorkovskii, M.A.; Tomilin, A.N. Post-Translational Modifications of Extracellular Proteasome. Molecules 2020, 25, 3504.
- Hanna, J.; Guerra-Moreno, A.; Ang, J.; Micoogullari, Y. Protein Degradation and the Pathologic Basis of Disease. Am. J. Pathol. 2019, 189, 94–103.
- Thibaudeau, T.A.; Smith, D.M. A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 2019, 71, 170–197.
- Rice, D.S.; Northcutt, G.M.; Kurschner, C. The Lnx Family Proteins Function as Molecular Scaffolds for Numb Family Proteins. Mol. Cell Neurosci. 2001, 18, 525–540.
- Xie, Y.; Zhao, W.; Wang, W.; Zhao, S.; Tang, R.; Ying, K.; Zhou, Z.; Mao, Y. Identification of a human LNX protein containing multiple PDZ domains. Biochem. Genet. 2001, 39, 117–126.
- Katoh, M.; Katoh, M. Identification and characterization of PDZRN3 and PDZRN4 genes in silico. Int. J. Mol. Med. 2004, 13, 607–613.
- Katoh, M.; Katoh, M. Identification and characterization of human PDZRN4L gene and mouse Pdzrn4l gene in silico. Int. J. Mol. Med. 2004, 13, 923–927.
- Romero, G.; Von Zastrow, M.; Friedman, P.A. Role of PDZ Proteins in Regulating Trafficking, Signaling, and Function of GPCRs: Means, Motif, and Opportunity. Adv. Pharm. 2011, 62, 279–314.
- Wolting, C.D.; Griffiths, E.K.; Sarao, R.; Prevost, B.C.; Wybenga-Groot, L.E.; McGlade, C.J. Biochemical and Computational Analysis of LNX1 Interacting Proteins. PLoS ONE 2011, 6, e26248.
- Guo, Z.; Song, E.; Ma, S.; Wang, X.; Gao, S.; Shao, C.; Hu, S.; Jia, L.; Tian, R.; Xu, T.; et al. Proteomics Strategy to Identify Substrates of LNX, a PDZ Domain-containing E3 Ubiquitin Ligase. J. Proteome Res. 2012, 11, 4847–4862.
- Lenihan, J.A.; Saha, O.; Young, P. Proteomic analysis reveals novel ligands and substrates for LNX1 E3 ubiquitin ligase. PLoS ONE 2017, 12, e0187352.
- Young, P. LNX1/LNX2 proteins: Functions in neuronal signalling and beyond. Neuronal Signal. 2018, 2, NS20170191.
- Pece, S.; Confalonieri, S.; Romano, P.R.; Di Fiore, P.P. NUMB-ing down cancer by more than just a NOTCH. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2011, 1815, 26–43.
- Nie, J.; McGill, M.A.; Dermer, M.; Dho, S.E.; Wolting, C.D.; McGlade, C.J. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002, 21, 93–102.
- Nie, J.; Li, S.S.-C.; McGlade, C.J. A Novel PTB-PDZ Domain Interaction Mediates Isoform-specific Ubiquitylation of Mammalian Numb. J. Biol. Chem. 2004, 279, 20807–20815.
- Won, M.; Ro, H.; Dawid, I.B. Lnx2 ubiquitin ligase is essential for exocrine cell differentiation in the early zebrafish pancreas. Proc. Nat. Acad. Sci. USA 2015, 112, 12426–12431.
- Nayak, D.; Sivaraman, J. Structural basis for the indispensable role of a unique zinc finger motif in LNX2 ubiquitination. Oncotarget 2015, 6, 34342–34357.
- Hekstra, D.R.; White, K.I.; Socolich, M.A.; Henning, R.W.; Šrajer, R.W.H.V.; Ranganathan, R. Electric-field-stimulated protein mechanics. Nat. Cell Biol. 2016, 540, 400–405.
- Nayak, D.; Sivaraman, J. Structure of LNX1:Ubc13 ~ Ubiquitin Complex Reveals the Role of Additional Motifs for the E3 Ligase Activity of LNX1. J. Mol. Biol. 2018, 430, 1173–1188.
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1843, 47–60.
- Weiss, A.; Baumgartner, M.; Radziwill, G.; Dennler, J.; Moelling, K. c-Src is a PDZ interaction partner and substrate of the E3 ubiquitin ligase Ligand-of-Numb protein X1. FEBS Lett. 2007, 581, 5131–5136.
- Lazzari, E.; El-Halawany, M.; De March, M.; Valentino, F.; Cantatore, F.; Migliore, C.; Onesti, S.; Meroni, G. Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H. Cells 2019, 8, 254.
- Maitland, M.E.R.; Onea, G.; Chiasson, C.A.; Wang, X.; Ma, J.; Moor, S.E.; Barber, K.R.; Lajoie, G.A.; Shaw, G.S.; Schild-Poulter, C. The mammalian CTLH complex is an E3 ubiquitin ligase that targets its subunit muskelin for degradation. Sci. Rep. 2019, 9, 1–14.
- Lu, Z.; Je, H.-S.; Young, P.; Gross, J.; Lu, B.; Feng, G. Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J. Cell Biol. 2007, 177, 1077–1089.
- Ro, H.; Dawid, I.B. Organizer restriction through modulation of Bozozok stability by the E3 ubiquitin ligase Lnx-like. Nat. Cell Biol. 2009, 11, 1121–1127.
- Fu, B.; Xue, W.; Zhang, H.; Zhang, R.; Feldman, K.; Zhao, Q.; Zhang, S.; Shi, L.; Pavani, K.C.; Nian, W.; et al. MicroRNA-325-3p Facilitates Immune Escape of Mycobacterium tuberculosis through Targeting LNX1 via NEK6 Accumulation to Promote Anti-Apoptotic STAT3 Signaling. mBio 2020, 11.
- Lee, C.-H.; Yang, J.-R.; Chen, C.-Y.; Tsai, M.-H.; Hung, P.-F.; Chen, S.-J.; Chiang, S.-L.; Chang, H.; Lin, P. Novel STAT3 Inhibitor LDOC1 Targets Phospho-JAK2 for Degradation by Interacting with LNX1 and Regulates the Aggressiveness of Lung Cancer. Cancers 2019, 11, 63.
- Lee, A.K.; Potts, P.R. A Comprehensive Guide to the MAGE Family of Ubiquitin Ligases. J. Mol. Biol. 2017, 429, 1114–1142.
- Sang, M.; Wang, L.; Ding, C.; Zhou, X.; Wang, B.; Wang, L.; Lian, Y.; Shan, B. Melanoma-associated antigen genes-an update. Cancer Lett 2011, 302, 85–90.
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING Protein Complexes Comprise a Family of E3 Ubiquitin Ligases. Mol. Cell 2010, 39, 963–974.
- Kozakova, L.; Vondrova, L.; Stejskal, K.; Charalabous, P.; Kolesar, P.; Lehmann, A.R.; Uldrijan, S.; Sanderson, C.M.; Zdrahal, Z.; Palecek, J.J. The melanoma-associated antigen 1 (MAGEA1) protein stimulates the E3 ubiquitin-ligase activity of TRIM31 within a TRIM31-MAGEA1-NSE4 complex. Cell Cycle 2015, 14, 920–930.
- Park, R.; Kim, H.; Jang, M.; Jo, D.; Park, Y.-I.; Namkoong, S.; Lee, J.I.; Jang, I.-S.; Park, J. LNX1 contributes to tumor growth by down-regulating p53 stability. FASEB J. 2019, 33, 13216–13227.
- Kansaku, A.; Hirabayashi, S.; Mori, H.; Fujiwara, N.; Kawata, A.; Ikeda, M.; Rokukawa, C.; Kurihara, H.; Hata, Y. Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene 2006, 25, 5071–5084.
- Takahashi, S.; Iwamoto, N.; Sasaki, H.; Ohashi, M.; Oda, Y.; Tsukita, S.; Furuse, M. The E3 ubiquitin ligase LNX1p80 promotes the removal of claudins from tight junctions in MDCK cells. J. Cell Sci. 2009, 122, 985–994.
- Tsukita, S.; Tanaka, H.; Tamura, A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem. Sci. 2019, 44, 141–152.
- Sollerbrant, K.; Raschperger, E.; Mirza, M.; Engström, U.; Philipson, L.; Ljungdahl, P.O.; Pettersson, R.F. The Coxsackievirus and Adenovirus Receptor (CAR) Forms a Complex with the PDZ Domain-containing Protein Ligand-of-Numb Protein-X (LNX). J. Biol. Chem. 2002, 278, 7439–7444.
- Mirza, M.; Raschperger, E.; Philipson, L.; Pettersson, R.F.; Sollerbrant, K. The cell surface protein coxsackie- and adenovirus receptor (CAR) directly associates with the Ligand-of-Numb Protein-X2 (LNX2). Exp. Cell Res. 2005, 309, 110–120.
- Mirza, M.; Hreinsson, J.; Strand, M.-L.; Hovatta, O.; Söder, O.; Philipson, L.; Pettersson, R.F.; Sollerbrant, K. Coxsackievirus and adenovirus receptor (CAR) is expressed in male germ cells and forms a complex with the differentiation factor JAM-C in mouse testis. Exp. Cell Res. 2006, 312, 817–830.
- Goodenough, D.A.; Paul, D.L. Gap Junctions. Cold Spring Harb. Perspect. Biol. 2009, 1, a002576.
- Skerrett, I.M.; Williams, J.B. A structural and functional comparison of gap junction channels composed of connexins and innexins. Dev. Neurobiol. 2017, 77, 522–547.
- Yamasaki, R. Connexins in health and disease. Clin. Exp. Neuroimmunol. 2018, 9, 30–36.
- Nagy, J.I.; Pereda, A.E.; Rash, J.E. Electrical synapses in mammalian CNS: Past eras, present focus and future directions. Biochim. et Biophys. Acta (BBA)-Biomembr. 2018, 1860, 102–123.
- Laing, J.G.; Tadros, P.N.; Westphale, E.M.; Beyer, E.C. Degradation of Connexin43 Gap Junctions Involves both the Proteasome and the Lysosome. Exp. Cell Res. 1997, 236, 482–492.
- Falk, M.M.; Kells, R.M.; Berthoud, V.M. Degradation of connexins and gap junctions. FEBS Lett. 2014, 588, 1221–1229.
- Lynn, B.D.; Li, X.; Hormuzdi, S.G.; Griffiths, E.K.; McGlade, C.J.; Nagy, J.I. E3 ubiquitin ligases LNX 1 and LNX 2 localize at neuronal gap junctions formed by connexin36 in rodent brain and molecularly interact with connexin36. Eur. J. Neurosci. 2018, 48, 3062–3081.
- Fujita, Y.; Krause, G.; Scheffner, M.; Zechner, D.; Leddy, H.E.M.; Behrens, J.; Sommer, T.; Birchmeier, W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002, 4, 222–231.
- Traweger, A.; Fang, D.; Liu, Y.-C.; Stelzhammer, W.; Krizbai, I.A.; Fresser, F.; Bauer, H.-C.; Bauer, H. The Tight Junction-specific Protein Occludin Is a Functional Target of the E3 Ubiquitin-protein Ligase Itch. J. Biol. Chem. 2002, 277, 10201–10208.
- Kaabeche, K.; Guenou, H.; Bouvard, D.; Didelot, N.; Listrat, A.; Marie, P.J. Cbl-mediated ubiquitination of 5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J. Cell Sci. 2005, 118, 1223–1232.
- Knights, A.J.; Funnell, A.P.W.; Crossley, M.; Pearson, R.C. Holding Tight: Cell Junctions and Cancer Spread. Trends Cancer Res. 2012, 8, 61–69.
- D’Agostino, M.; Tornillo, G.; Caporaso, M.G.; Barone, M.V.; Ghigo, E.; Bonatti, S.; Mottola, G. Ligand of Numb proteins LNX1p80 and LNX2 interact with the human glycoprotein CD8α and promote its ubiquitylation and endocytosis. J. Cell Sci 2011, 124, 3545–3556.
- Liu, X.-D.; Zhu, X.-N.; Halford, M.M.; Xu, T.-L.; Henkemeyer, M.; Xu, N.-J. Retrograde regulation of mossy fiber axon targeting and terminal maturation via postsynaptic Lnx1. J. Cell Biol. 2018, 217, 4007–4024.
- Liu, X.-D.; Ai, P.-H.; Zhu, X.-N.; Pan, Y.-B.; Halford, M.M.; Henkemeyer, M.; Feng, D.-F.; Xu, T.-L.; Sun, S.; Xu, N.-J. Hippocampal Lnx1–NMDAR multiprotein complex mediates initial social memory. Mol. Psychiatry 2019, 1–14.
- Rocha-Muñoz, A.; Núñez, E.; Arribas-González, E.; López-Corcuera, B.; Aragón, C.; de Juan-Sanz, J. E3 ubiquitin ligases LNX1 and LNX2 are major regulators of the presynaptic glycine transporter GlyT2. Sci Rep. 2019, 9, 14944.
References
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle’s Dynamics and Signaling. PLoS ONE 2008, 3, e1487.
- Clague, M.J.; Heride, C.; Urbé, S. The demographics of the ubiquitin system. Trends Cell Biol. 2015, 25, 417–426.
- Lipkowitz, S.; Weissman, A.M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 2011, 11, 629–643.
- Ro, H.; Hur, T.-L.; Rhee, M. Ubiquitin conjugation system for body axes specification in vertebrates. Anim. Cells Syst. 2015, 19, 87–95.
- Bhat, K.P.; Greer, S.F. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2011, 1809, 150–155.
- Tsimokha, A.S.; Artamonova, T.O.; Diakonov, E.E.; Khodorkovskii, M.A.; Tomilin, A.N. Post-Translational Modifications of Extracellular Proteasome. Molecules 2020, 25, 3504.
- Hanna, J.; Guerra-Moreno, A.; Ang, J.; Micoogullari, Y. Protein Degradation and the Pathologic Basis of Disease. Am. J. Pathol. 2019, 189, 94–103.
- Thibaudeau, T.A.; Smith, D.M. A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 2019, 71, 170–197.
- Rice, D.S.; Northcutt, G.M.; Kurschner, C. The Lnx Family Proteins Function as Molecular Scaffolds for Numb Family Proteins. Mol. Cell Neurosci. 2001, 18, 525–540.
- Xie, Y.; Zhao, W.; Wang, W.; Zhao, S.; Tang, R.; Ying, K.; Zhou, Z.; Mao, Y. Identification of a human LNX protein containing multiple PDZ domains. Biochem. Genet. 2001, 39, 117–126.
- Katoh, M.; Katoh, M. Identification and characterization of PDZRN3 and PDZRN4 genes in silico. Int. J. Mol. Med. 2004, 13, 607–613.
- Katoh, M.; Katoh, M. Identification and characterization of human PDZRN4L gene and mouse Pdzrn4l gene in silico. Int. J. Mol. Med. 2004, 13, 923–927.
- Romero, G.; Von Zastrow, M.; Friedman, P.A. Role of PDZ Proteins in Regulating Trafficking, Signaling, and Function of GPCRs: Means, Motif, and Opportunity. Adv. Pharm. 2011, 62, 279–314.
- Wolting, C.D.; Griffiths, E.K.; Sarao, R.; Prevost, B.C.; Wybenga-Groot, L.E.; McGlade, C.J. Biochemical and Computational Analysis of LNX1 Interacting Proteins. PLoS ONE 2011, 6, e26248.
- Guo, Z.; Song, E.; Ma, S.; Wang, X.; Gao, S.; Shao, C.; Hu, S.; Jia, L.; Tian, R.; Xu, T.; et al. Proteomics Strategy to Identify Substrates of LNX, a PDZ Domain-containing E3 Ubiquitin Ligase. J. Proteome Res. 2012, 11, 4847–4862.
- Lenihan, J.A.; Saha, O.; Young, P. Proteomic analysis reveals novel ligands and substrates for LNX1 E3 ubiquitin ligase. PLoS ONE 2017, 12, e0187352.
- Young, P. LNX1/LNX2 proteins: Functions in neuronal signalling and beyond. Neuronal Signal. 2018, 2, NS20170191.
- Pece, S.; Confalonieri, S.; Romano, P.R.; Di Fiore, P.P. NUMB-ing down cancer by more than just a NOTCH. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2011, 1815, 26–43.
- Nie, J.; McGill, M.A.; Dermer, M.; Dho, S.E.; Wolting, C.D.; McGlade, C.J. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002, 21, 93–102.
- Nie, J.; Li, S.S.-C.; McGlade, C.J. A Novel PTB-PDZ Domain Interaction Mediates Isoform-specific Ubiquitylation of Mammalian Numb. J. Biol. Chem. 2004, 279, 20807–20815.
- Won, M.; Ro, H.; Dawid, I.B. Lnx2 ubiquitin ligase is essential for exocrine cell differentiation in the early zebrafish pancreas. Proc. Nat. Acad. Sci. USA 2015, 112, 12426–12431.
- Nayak, D.; Sivaraman, J. Structural basis for the indispensable role of a unique zinc finger motif in LNX2 ubiquitination. Oncotarget 2015, 6, 34342–34357.
- Hekstra, D.R.; White, K.I.; Socolich, M.A.; Henning, R.W.; Šrajer, R.W.H.V.; Ranganathan, R. Electric-field-stimulated protein mechanics. Nat. Cell Biol. 2016, 540, 400–405.
- Nayak, D.; Sivaraman, J. Structure of LNX1:Ubc13 ~ Ubiquitin Complex Reveals the Role of Additional Motifs for the E3 Ligase Activity of LNX1. J. Mol. Biol. 2018, 430, 1173–1188.
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1843, 47–60.
- Weiss, A.; Baumgartner, M.; Radziwill, G.; Dennler, J.; Moelling, K. c-Src is a PDZ interaction partner and substrate of the E3 ubiquitin ligase Ligand-of-Numb protein X1. FEBS Lett. 2007, 581, 5131–5136.
- Lazzari, E.; El-Halawany, M.; De March, M.; Valentino, F.; Cantatore, F.; Migliore, C.; Onesti, S.; Meroni, G. Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H. Cells 2019, 8, 254.
- Maitland, M.E.R.; Onea, G.; Chiasson, C.A.; Wang, X.; Ma, J.; Moor, S.E.; Barber, K.R.; Lajoie, G.A.; Shaw, G.S.; Schild-Poulter, C. The mammalian CTLH complex is an E3 ubiquitin ligase that targets its subunit muskelin for degradation. Sci. Rep. 2019, 9, 1–14.
- Lu, Z.; Je, H.-S.; Young, P.; Gross, J.; Lu, B.; Feng, G. Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J. Cell Biol. 2007, 177, 1077–1089.
- Ro, H.; Dawid, I.B. Organizer restriction through modulation of Bozozok stability by the E3 ubiquitin ligase Lnx-like. Nat. Cell Biol. 2009, 11, 1121–1127.
- Fu, B.; Xue, W.; Zhang, H.; Zhang, R.; Feldman, K.; Zhao, Q.; Zhang, S.; Shi, L.; Pavani, K.C.; Nian, W.; et al. MicroRNA-325-3p Facilitates Immune Escape of Mycobacterium tuberculosis through Targeting LNX1 via NEK6 Accumulation to Promote Anti-Apoptotic STAT3 Signaling. mBio 2020, 11.
- Lee, C.-H.; Yang, J.-R.; Chen, C.-Y.; Tsai, M.-H.; Hung, P.-F.; Chen, S.-J.; Chiang, S.-L.; Chang, H.; Lin, P. Novel STAT3 Inhibitor LDOC1 Targets Phospho-JAK2 for Degradation by Interacting with LNX1 and Regulates the Aggressiveness of Lung Cancer. Cancers 2019, 11, 63.
- Lee, A.K.; Potts, P.R. A Comprehensive Guide to the MAGE Family of Ubiquitin Ligases. J. Mol. Biol. 2017, 429, 1114–1142.
- Sang, M.; Wang, L.; Ding, C.; Zhou, X.; Wang, B.; Wang, L.; Lian, Y.; Shan, B. Melanoma-associated antigen genes-an update. Cancer Lett 2011, 302, 85–90.
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING Protein Complexes Comprise a Family of E3 Ubiquitin Ligases. Mol. Cell 2010, 39, 963–974.
- Kozakova, L.; Vondrova, L.; Stejskal, K.; Charalabous, P.; Kolesar, P.; Lehmann, A.R.; Uldrijan, S.; Sanderson, C.M.; Zdrahal, Z.; Palecek, J.J. The melanoma-associated antigen 1 (MAGEA1) protein stimulates the E3 ubiquitin-ligase activity of TRIM31 within a TRIM31-MAGEA1-NSE4 complex. Cell Cycle 2015, 14, 920–930.
- Park, R.; Kim, H.; Jang, M.; Jo, D.; Park, Y.-I.; Namkoong, S.; Lee, J.I.; Jang, I.-S.; Park, J. LNX1 contributes to tumor growth by down-regulating p53 stability. FASEB J. 2019, 33, 13216–13227.
- Kansaku, A.; Hirabayashi, S.; Mori, H.; Fujiwara, N.; Kawata, A.; Ikeda, M.; Rokukawa, C.; Kurihara, H.; Hata, Y. Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene 2006, 25, 5071–5084.
- Takahashi, S.; Iwamoto, N.; Sasaki, H.; Ohashi, M.; Oda, Y.; Tsukita, S.; Furuse, M. The E3 ubiquitin ligase LNX1p80 promotes the removal of claudins from tight junctions in MDCK cells. J. Cell Sci. 2009, 122, 985–994.
- Tsukita, S.; Tanaka, H.; Tamura, A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem. Sci. 2019, 44, 141–152.
- Sollerbrant, K.; Raschperger, E.; Mirza, M.; Engström, U.; Philipson, L.; Ljungdahl, P.O.; Pettersson, R.F. The Coxsackievirus and Adenovirus Receptor (CAR) Forms a Complex with the PDZ Domain-containing Protein Ligand-of-Numb Protein-X (LNX). J. Biol. Chem. 2002, 278, 7439–7444.
- Mirza, M.; Raschperger, E.; Philipson, L.; Pettersson, R.F.; Sollerbrant, K. The cell surface protein coxsackie- and adenovirus receptor (CAR) directly associates with the Ligand-of-Numb Protein-X2 (LNX2). Exp. Cell Res. 2005, 309, 110–120.
- Mirza, M.; Hreinsson, J.; Strand, M.-L.; Hovatta, O.; Söder, O.; Philipson, L.; Pettersson, R.F.; Sollerbrant, K. Coxsackievirus and adenovirus receptor (CAR) is expressed in male germ cells and forms a complex with the differentiation factor JAM-C in mouse testis. Exp. Cell Res. 2006, 312, 817–830.
- Goodenough, D.A.; Paul, D.L. Gap Junctions. Cold Spring Harb. Perspect. Biol. 2009, 1, a002576.
- Skerrett, I.M.; Williams, J.B. A structural and functional comparison of gap junction channels composed of connexins and innexins. Dev. Neurobiol. 2017, 77, 522–547.
- Yamasaki, R. Connexins in health and disease. Clin. Exp. Neuroimmunol. 2018, 9, 30–36.
- Nagy, J.I.; Pereda, A.E.; Rash, J.E. Electrical synapses in mammalian CNS: Past eras, present focus and future directions. Biochim. et Biophys. Acta (BBA)-Biomembr. 2018, 1860, 102–123.
- Laing, J.G.; Tadros, P.N.; Westphale, E.M.; Beyer, E.C. Degradation of Connexin43 Gap Junctions Involves both the Proteasome and the Lysosome. Exp. Cell Res. 1997, 236, 482–492.
- Falk, M.M.; Kells, R.M.; Berthoud, V.M. Degradation of connexins and gap junctions. FEBS Lett. 2014, 588, 1221–1229.
- Lynn, B.D.; Li, X.; Hormuzdi, S.G.; Griffiths, E.K.; McGlade, C.J.; Nagy, J.I. E3 ubiquitin ligases LNX 1 and LNX 2 localize at neuronal gap junctions formed by connexin36 in rodent brain and molecularly interact with connexin36. Eur. J. Neurosci. 2018, 48, 3062–3081.
- Fujita, Y.; Krause, G.; Scheffner, M.; Zechner, D.; Leddy, H.E.M.; Behrens, J.; Sommer, T.; Birchmeier, W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002, 4, 222–231.
- Traweger, A.; Fang, D.; Liu, Y.-C.; Stelzhammer, W.; Krizbai, I.A.; Fresser, F.; Bauer, H.-C.; Bauer, H. The Tight Junction-specific Protein Occludin Is a Functional Target of the E3 Ubiquitin-protein Ligase Itch. J. Biol. Chem. 2002, 277, 10201–10208.
- Kaabeche, K.; Guenou, H.; Bouvard, D.; Didelot, N.; Listrat, A.; Marie, P.J. Cbl-mediated ubiquitination of 5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J. Cell Sci. 2005, 118, 1223–1232.
- Knights, A.J.; Funnell, A.P.W.; Crossley, M.; Pearson, R.C. Holding Tight: Cell Junctions and Cancer Spread. Trends Cancer Res. 2012, 8, 61–69.
- D’Agostino, M.; Tornillo, G.; Caporaso, M.G.; Barone, M.V.; Ghigo, E.; Bonatti, S.; Mottola, G. Ligand of Numb proteins LNX1p80 and LNX2 interact with the human glycoprotein CD8α and promote its ubiquitylation and endocytosis. J. Cell Sci 2011, 124, 3545–3556.
- Liu, X.-D.; Zhu, X.-N.; Halford, M.M.; Xu, T.-L.; Henkemeyer, M.; Xu, N.-J. Retrograde regulation of mossy fiber axon targeting and terminal maturation via postsynaptic Lnx1. J. Cell Biol. 2018, 217, 4007–4024.
- Liu, X.-D.; Ai, P.-H.; Zhu, X.-N.; Pan, Y.-B.; Halford, M.M.; Henkemeyer, M.; Feng, D.-F.; Xu, T.-L.; Sun, S.; Xu, N.-J. Hippocampal Lnx1–NMDAR multiprotein complex mediates initial social memory. Mol. Psychiatry 2019, 1–14.
- Rocha-Muñoz, A.; Núñez, E.; Arribas-González, E.; López-Corcuera, B.; Aragón, C.; de Juan-Sanz, J. E3 ubiquitin ligases LNX1 and LNX2 are major regulators of the presynaptic glycine transporter GlyT2. Sci Rep. 2019, 9, 14944.
