The first antibiotic-producing actinomycete (Streptomyces antibioticus) was described by Waksman and Woodruff in 1940. This discovery initiated the “actinomycetes era”, in which several species were identified and demonstrated to be a great source of bioactive compounds. However, the remarkable group of microorganisms and their potential for the production of bioactive agents were only partially exploited. This is caused by the fact that the growth of many actinomycetes cannot be reproduced on artificial media at laboratory conditions. In addition, sequencing, genome mining and bioactivity screening disclosed that numerous biosynthetic gene clusters (BGCs), encoded in actinomycetes genomes are not expressed and thus, the respective potential products remain uncharacterized. Therefore, a lot of effort was put into the development of technologies that facilitate the access to actinomycetes genomes and activation of their biosynthetic pathways. In this review (Antibiotics 2020, 9(8), 494; https://doi.org/10.3390/antibiotics9080494), we mainly focus on molecular tools and methods for genetic engineering of actinomycetes that have emerged in the field in the past five years (2015–2020). In addition, we highlight examples of successful application of the recently developed technologies in genetic engineering of actinomycetes for activation and/or improvement of the biosynthesis of secondary metabolites.
- actinomycetes
- natural products
- genetic engineering
1. Introduction
Actinomycetes are Gram-positive, mostly aerobic, filamentous bacteria of the phylum Actinobacteria [1][2][3][4][1–4]. They were initially regarded as organisms, which share morphological characteristics with both bacteria and fungi [5][6][5,6] and thus, they were considered as transitional forms between the two groups. This is reflected in the name “Actinomycetes” that was derived from Greek “atkis” or “aktin” (means ray) and “mykes” (means fungus) [7][8][7,8]. Actinomycetes have adapted to different ecological niches: terrestrial [9][10][11][12][13][14][15][16][17][18][9–18], aquatic [19][20][21][22][23][24][25][19–25] and artificial (“manmade”) [26][27][26,27]. In many cases, their biological function is indispensable. Some species are involved in decomposing complex mixtures of polymers in dead plants, animals and fungal materials contributing to humus formation and the recycling of biomaterials [28][29][28,29]. Actinomycetes can also provide a nitrogen-fixing function in symbiotic associations with plants [30][31][30,31] or take part in other complex, ecologically relevant interactions [32][33][34][32–34]. The most known example includes interactions with insects, where the antibiotics produced by actinomycetes protect the associated organism (e.g., termites, ants, beetles, wasps or they larvae) against detrimental microorganisms [35][36][37][38][39][40][35–40].
However, many researchers have drawn attention to actinomycetes mainly because of their biosynthetic potential for the production of secondary metabolites [41][42][43][41–43]. The discovery of the first actinomycete-derived antibiotic (actinomycin) [44][44] boosted the isolation of actinomycetes from diverse sources and the traditional screening involving the cultivation, extraction of metabolites and disk diffusion-based activity assays and/or chromatographic analysis [43][45][46][47][43,45–47]. Since the “Golden Age” of antibiotic discovery (1940s–1960s), several antimicrobial and other valuable compounds were discovered from actinomycetes. Many of these molecules have been developed to commercial products, including agrochemicals and pharmaceuticals [14][47][48][49][50][51][14,47–51]. Thus, actinomycetes are one of the most prolific and important sources of bioactive compounds. Since the whole genome sequencing of the first actinomycetes, which was the genome of Streptomyces coelicolor A3 [52][52], hundreds actinomycetes genomes were sequenced and annotated. These data revealed that the genomes encode several biosynthetic gene clusters (BGCs) for the production of diverse secondary metabolites. However, only a few of the BGCs are expressed at standard laboratory conditions and, therefore, the biosynthetic potential of many actinomycetes remains unexploited [53][54][55][56][57][58][59][60][53–60]. Numerous strategies were developed to induce the expression of the BGCs and access the corresponding products.
2. Genetic Engineering of Actinomycetes
The currently used genetic engineering strategies for activation of BGC-expression and production of the respective compounds in actinomycetes include the expression of multiple copies of the whole BGC or factors that are limiting the production, expression of activator genes, deletion of genes encoding repressors of the BGC, refactoring of the BGC by substitution or modification of native regulatory elements (e.g., promoters) and/or expression of the BGC in optimized (e.g., genome-minimized, precursor-optimized), native or heterologous hosts [61][62][63][64][65][69–73] (Figure 1 and Figure 2). These in vivo engineering approaches require genetic manipulation that depends on molecular tools for assembling of genetic constructs and methods for their introduction into the host. In the past four decades, several barriers were identified that retarded the process of natural product discovery from actinomycetes. Lack of compatible molecular tools, limited cloning and DNA transfer methods, DNA degradation, genetic instability, high guanine-cytosine content (GC-content) of the genomes, and different codon usage of the newly introduced foreign DNA are barriers that often prevent the genetic manipulation of actinomycetes (Figure 1).
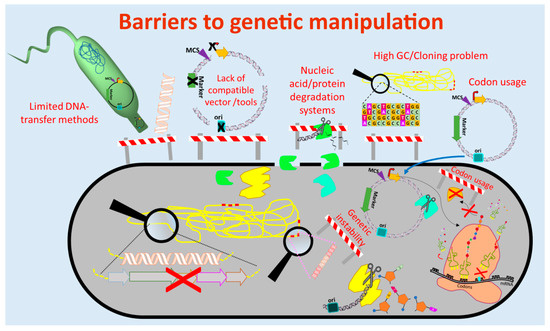
Figure 1. Barriers to genetic manipulation of actinomycetes. Obstacles hindering the genetic manipulation and engineering of actinomycetes include the lack of strain-compatible tools, cloning methods for high guanine-cytosine content (GC-content) DNA sequences as well as transfer methods for introduction of the genetic constructs into the host. Furthermore, nucleic acid degradation systems present in actinomycetes and differences in codon usage may result in fragmentation of the genetic constructs and production of non-functional proteins, respectively. (Single elements of the figure (e.g., ribosome) were re-used from a previous publication [66][74] with permission from the Royal Society of Chemistry.)
Therefore, enormous efforts have been undertaken to develop tools and methods to overcome these obstacles (Figure 2). Advances in sequencing technics [67][75] and bioinformatics tools for genome mining [68][63] facilitate cost-effective sequencing and a fast identification of BGCs and other potential targets for genetic engineering. Molecular biology enzymes [69][70][71][76–78], fast cloning strategies and synthetic biology which involves vectors and genetic parts (e.g., attachment sites, replicons, selection markers, promoters, terminators) [72][73][74][75][76][77][79–84] were optimized for introduction and maintenance of additional or new genetic material in actinomycetes strains. Methods for transfer of the generated construct into the cell (e.g., protoplast transformation, conjugation) were established [66][78][74,85].
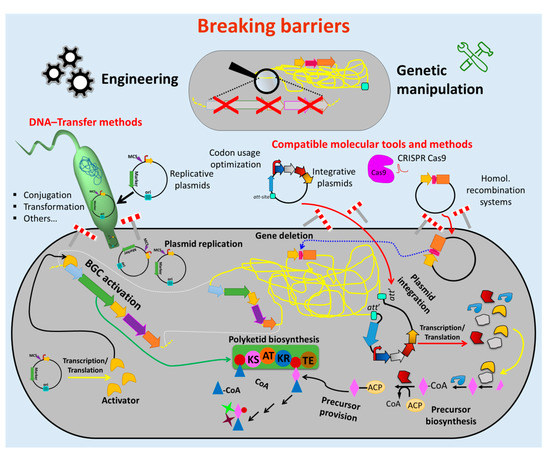
Figure 2. Breaking down the barriers to genetic manipulation and engineering of actinomycetes. Numerous methods and tools were developed to facilitate the genetic manipulation of actinomycetes. Suitable vectors and other vehicles (e.g., integrative and replicative systems) as well as fast and easy cloning strategies were developed to assemble genetic constructs, which are introduced into the host by using different transfer methods (e.g., conjugation and protoplast transformation). Protocols for avoiding DNA-degradation (e.g., treatment of the DNA with NaOH) and solutions for ensuring correct transcription/translation (e.g., codon usage optimization) were established. These innovations led to the generation of many gene deletions and (over)expression mutants (e.g., deletion of competitive biosynthetic pathways, overexpression of activators for activation of silent biosynthetic gene cluster (BGC)). This enormous progress facilitated the metabolic engineering of actinomycetes chassis for manufacturing bioactive natural products, including antibiotics. (Single elements of the figure (e.g., schematic representation of plasmid backbone) were re-used from a previous publication [66][74] with permission from the Royal Society of Chemistry.).
During the past five years (2015–2020) a variety of technologies and protocols for engineering of actinomycetes genomes have been established. Therefore, we believe that a summary of the recent developments (Antibiotics 2020, 9(8), 494; https://doi.org/10.3390/antibiotics9080494)[1] will be a valuable reference for the community focusing on actinomycetes and their natural products. To this end, we present an update on the molecular tools and methods for genetic engineering of actinomycetes. In the first part, details on new cloning strategies were described. The second part contains an overview on recent advances in “genetic bricks” and molecular tools for genetic manipulation of actinomycetes. As there is tremendous progress in the development of cloning strategies and CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR associated)-based tools, these two sections have a “lexicon-like” structure. This should ensure an easy and time-saving search for the diverse tools and their explanation.
References
- Mitousis, Lena; Thoma, Yvonne; and Ewa M. Musiol-Kroll, Ewa M.; An Update on Molecular Tools for Genetic Engineering of Actinomycetes—The Source of Important Antibiotics and Other Valuable Compounds . Antibiotics 2020, 9, article number: 494, https://doi.org/10.3390/antibiotics9080494.Nouioui, I.; Carro, L.; Garcia-Lopez, M.; Meier-Kolthoff, J.P.; Woyke, T.; Kyrpides, N.C.; Pukall, R.; Klenk, H.P.; Goodfellow, M.; Goker, M. Genome-Based Taxonomic Classification of the Phylum Actinobacteria. Front. Microbiol. 2018, 9.
- Whitman, W.; Goodfellow, M.; Kämpfer, P.; Busse, H.-J.; Trujillo, M.; Ludwig, W.; Suzuki, K.-I.; Parte, A. Bergey’s Manual of Systematic Bacteriology, “the Actinobacteria”, 2nd ed.; Springer-Verlag: New York, NY, USA, 2012; pp. 1–2083.
- Gao, B.; Gupta, R.S. Phylogenetic Framework and Molecular Signatures for the Main Clades of the Phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 66–112.
- Verma, M.; Lal, D.; Kaur, J.; Saxena, A.; Kaur, J.; Anand, S.; Lal, R. Phylogenetic Analyses of Phylum Actinobacteria Based on Whole Genome Sequences. Res. Microbiol. 2013, 164, 718–728.
- Waksman, S.A. Studies in the Metabolism of Actinomycetes: Part II. J. Bacteriol. 1919, 4, 307–330.
- Waksman, S.A. Studies in the Metabolism of Actinomycetes: Part I. J. Bacteriol. 1919, 4, 189–216.
- Waksman, S.A. On the Classification of Actinomycetes. J. Bacteriol. 1940, 39, 549–558.
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; Van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43.
- Goodfellow, M.; Nouioui, I.; Sanderson, R.; Xie, F.; Bull, A.T. Rare Taxa and Dark Microbial Matter: Novel Bioactive Actinobacteria Abound in Atacama Desert Soils. Antonie Van Leeuwenhoek 2018, 111, 1315–1332.
- Singh, R.; Dubey, A.K. Diversity and Applications of Endophytic Actinobacteria of Plants in Special and Other Ecological Niches. Front. Microbiol. 2018, 9, 1767.
- Guo, X.; Liu, N.; Li, X.; Ding, Y.; Shang, F.; Gao, Y.; Ruan, J.; Huang, Y. Red Soils Harbor Diverse Culturable Actinomycetes That Are Promising Sources of Novel Secondary Metabolites. Appl. Environ. Microbiol. 2015, 81, 3086–3103.
- Jackson, T.A. Fossil Actinomycetes in Middle Precambrian Glacial Varves. Science 1967, 155, 1003–1005.
- Poomthongdee, N.; Duangmal, K.; Pathom-aree, W. Acidophilic Actinomycetes from Rhizosphere Soil: Diversity and Properties Beneficial to Plants. J. Antibiot. (Tokyo) 2015, 68, 106–114.
- Matsumoto, A.; Takahashi, Y. Endophytic Actinomycetes: Promising Source of Novel Bioactive Compounds. J. Antibiot. (Tokyo) 2017, 70, 514–519.
- Chouvenc, T.; Elliott, M.L.; Sobotnik, J.; Efstathion, C.A.; Su, N.Y. The Termite Fecal Nest: A Framework for the Opportunistic Acquisition of Beneficial Soil Streptomyces (Actinomycetales: Streptomycetaceae). Environ. Entomol. 2018, 47, 1431–1439.
- Scott, J.J.; Oh, D.C.; Yuceer, M.C.; Klepzig, K.D.; Clardy, J.; Currie, C.R. Bacterial Protection of Beetle-Fungus Mutualism. Science 2008, 322, 63.
- Zaher, F.; Isenberg, H.D.; Rosenfeld, M.H.; Schatz, A. The Distribution of Soil Actinomycetes Antagonistic to Protozoa. J. Parasitol. 1953, 39, 33–37.
- Takasuka, T.E.; Book, A.J.; Lewin, G.R.; Currie, C.R.; Fox, B.G. Aerobic Deconstruction of Cellulosic Biomass by an Insect-Associated Streptomyces. Sci. Rep. 2013, 3, 1030.
- Yang, T.; Yamada, K.; Zhou, T.; Harunari, E.; Igarashi, Y.; Terahara, T.; Kobayashi, T.; Imada, C. Akazamicin, a Cytotoxic Aromatic Polyketide from Marine-Derived Nonomuraea Sp. J. Antibiot. (Tokyo) 2019, 72, 202–209.
- Iniyan, A.M.; Sudarman, E.; Wink, J.; Kannan, R.R.; Vincent, S.G.P. Ala-Geninthiocin, a New Broad Spectrum Thiopeptide Antibiotic, Produced by a Marine Streptomyces Sp. Icn19. J. Antibiot. (Tokyo) 2019, 72, 99–105.
- Jiang, C.; Xu, L. Diversity of Aquatic Actinomycetes in Lakes of the Middle Plateau, Yunnan, China. Appl. Environ. Microbiol. 1996, 62, 249–253.
- Lee, G.C.; Kim, Y.S.; Kim, M.J.; Oh, S.A.; Choi, I.; Choi, J.; Park, J.G.; Chong, C.K.; Kim, Y.Y.; Lee, K.; et al. Presence, Molecular Characteristics and Geosmin Producing Ability of Actinomycetes Isolated from South Korean Terrestrial and Aquatic Environments. Water Sci. Technol. 2011, 63, 2745–2751.
- Saadoun, I.; Hameed, K.M.; Moussauui, A. Characterization and Analysis of Antibiotic Activity of Some Aquatic Actinomycetes. Microbios. 1999, 99, 173–179.
- Ueda, J.Y.; Khan, S.T.; Takagi, M.; Shin-ya, K. Jbir-58, A New Salicylamide Derivative, Isolated from a Marine Sponge-Derived Streptomyces Sp. Spd081030me-02. J. Antibiot. (Tokyo) 2010, 63, 267–269.
- El-Hawary, S.S.; Sayed, A.M.; Mohammed, R.; Khanfar, M.A.; Rateb, M.E.; Mohammed, T.A.; Hajjar, D.; Hassan, H.M.; Gulder, T.A.M.; Abdelmohsen, U.R. New Pim-1 Kinase Inhibitor from the Co-Culture of Two Sponge-Associated Actinomycetes. Front. Chem. 2018, 6, 538.
- Mehbub, M.F.; Tanner, J.E.; Barnett, S.J.; Franco, C.M.; Zhang, W. The Role of Sponge-Bacteria Interactions: The Sponge Aplysilla rosea Challenged by Its Associated Bacterium Streptomyces act-52a in a Controlled Aquarium System. Appl. Microbiol. Biotechnol. 2016, 100, 10609–10626.
- Minas, W.; Bailey, J.E.; Duetz, W. Streptomycetes in Micro-Cultures: Growth, Production of Secondary Metabolites, and Storage and Retrieval in the 96-Well Format. Antonie Van Leeuwenhoek 2000, 78, 297–305.
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes Benefaction Role in Soil and Plant Health. Microb. Pathog. 2017, 111, 458–467.
- Saini, A.; Aggarwal, N.K.; Sharma, A.; Yadav, A. Actinomycetes: A Source of Lignocellulolytic Enzymes. Enzyme Res. 2015, 2015.
- Sellstedt, A.; Richau, K.H. Aspects of Nitrogen-Fixing Actinobacteria, in Particular Free-Living and Symbiotic Frankia. FEMS Microbiol. Lett. 2013, 342, 179–186.
- Dahal, B.; NandaKafle, G.; Perkins, L.; Brozel, V.S. Diversity of Free-Living Nitrogen Fixing Streptomyces in Soils of the Badlands of South Dakota. Microbiol. Res. 2017, 195, 31–39.
- Liu, C.; Jiang, Y.; Wang, X.; Chen, D.; Chen, X.; Wang, L.; Han, L.; Huang, X.; Jiang, C. Diversity, Antimicrobial Activity, and Biosynthetic Potential of Cultivable Actinomycetes Associated with Lichen Symbiosis. Microb. Ecol. 2017, 74, 570–584.
- Li, H.; Sosa-Calvo, J.; Horn, H.A.; Pupo, M.T.; Clardy, J.; Rabeling, C.; Schultz, T.R.; Currie, C.R. Convergent Evolution of Complex Structures for Ant-Bacterial Defensive Symbiosis in Fungus-Farming Ants. Proc. Natl. Acad. Sci. USA 2018, 115, 10720–10725.
- Van der Meij, A.; Willemse, J.; Schneijderberg, M.A.; Geurts, R.; Raaijmakers, J.M.; Van Wezel, G.P. Inter- and Intracellular Colonization of Arabidopsis Roots by Endophytic Actinobacteria and the Impact of Plant Hormones on Their Antimicrobial Activity. Antonie Van Leeuwenhoek 2018, 111, 679–690.
- Kaltenpoth, M.; Yildirim, E.; Gurbuz, M.F.; Herzner, G.; Strohm, E. Refining the Roots of the Beewolf-Streptomyces Symbiosis: Antennal Symbionts in the Rare Genus Philanthinus (Hymenoptera, Crabronidae). Appl. Environ. Microbiol. 2012, 78, 822–827.
- Kaltenpoth, M. Actinobacteria as Mutualists: General Healthcare for Insects? Trends Microbiol. 2009, 17, 529–535.
- Kurtboke, D.I.; French, J.R.; Hayes, R.A.; Quinn, R.J. Eco-Taxonomic Insights into Actinomycete Symbionts of Termites for Discovery of Novel Bioactive Compounds. Adv. Biochem. Eng. Biotechnol. 2015, 147, 111–135.
- Visser, A.A.; Nobre, T.; Currie, C.R.; Aanen, D.K.; Poulsen, M. Exploring the Potential for Actinobacteria as Defensive Symbionts in Fungus-Growing Termites. Microb. Ecol. 2012, 63, 975–985.
- Kim, K.H.; Ramadhar, T.R.; Beemelmanns, C.; Cao, S.; Poulsen, M.; Currie, C.R.; Clardy, J. Natalamycin A, an Ansamycin from a Termite-Associated Streptomyces Sp. Chem. Sci. 2014, 5, 4333–4338.
- Beemelmanns, C.; Ramadhar, T.R.; Kim, K.H.; Klassen, J.L.; Cao, S.; Wyche, T.P.; Hou, Y.; Poulsen, M.; Bugni, T.S.; Currie, C.R.; et al. Macrotermycins A-D, Glycosylated Macrolactams from a Termite-Associated Amycolatopsis Sp. M39. Org. Lett. 2017, 19, 1000–1003.
- Robertsen, H.L.; Musiol-Kroll, E.M. Actinomycete-Derived Polyketides as a Source of Antibiotics and Lead Structures for the Development of New Antimicrobial Drugs. Antibiotics (Basel) 2019, 8, 157.
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249.
- Genilloud, O. Actinomycetes: Still a Source of Novel Antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232.
- Waksman, S.A.; Woodruff, H.B. Actinomyces antibioticus, a New Soil Organism Antagonistic to Pathogenic and Non-Pathogenic Bacteria. J. Bacteriol. 1941, 42, 231–249.
- Hug, J.J.; Bader, C.D.; Remskar, M.; Cirnski, K.; Muller, R. Concepts and Methods to Access Novel Antibiotics from Actinomycetes. Antibiotics (Basel) 2018, 7, 44.
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45.
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; Van Wezel, G.P. Chemical Ecology of Antibiotic Production by Actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416.
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic Basis for Natural Product Biosynthetic Diversity in the Actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384.
- Takahashi, Y.; Nakashima, T. Actinomycetes, an Inexhaustible Source of Naturally Occurring Antibiotics. Antibiotics (Basel) 2018, 7, 45.
- Landwehr, W.; Wolf, C.; Wink, J. Actinobacteria and Myxobacteria-Two of the Most Important Bacterial Resources for Novel Antibiotics. Curr. Top. Microbiol. Immunol. 2016, 398, 273–302.
- Dhakal, D.; Pokhrel, A.R.; Shrestha, B.; Sohng, J.K. Marine Rare Actinobacteria: Isolation, Characterization, and Strategies for Harnessing Bioactive Compounds. Front. Microbiol. 2017, 8, 1106.
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete Genome Sequence of the Model Actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147.
- Baral, B.; Akhgari, A.; Metsa-Ketela, M. Activation of Microbial Secondary Metabolic Pathways: Avenues and Challenges. Synth. Syst. Biotechnol. 2018, 3, 163–178.
- Onaka, H. Novel Antibiotic Screening Methods to Awaken Silent or Cryptic Secondary Metabolic Pathways in Actinomycetes. J. Antibiot. (Tokyo) 2017, 70, 865–870.
- Ochi, K.; Hosaka, T. New Strategies for Drug Discovery: Activation of Silent or Weakly Expressed Microbial Gene Clusters. Appl. Microbiol. Biotechnol. 2013, 97, 87–98.
- Becerril, A.; Alvarez, S.; Brana, A.F.; Rico, S.; Diaz, M.; Santamaria, R.I.; Salas, J.A.; Mendez, C. Uncovering Production of Specialized Metabolites by Streptomyces argillaceus: Activation of Cryptic Biosynthesis Gene Clusters Using Nutritional and Genetic Approaches. PLoS One 2018, 13, e0198145.
- Liu, Y.; Wei, W.P.; Ye, B.C. High GC Content Cas9-Mediated Genome-Editing and Biosynthetic Gene Cluster Activation in Saccharopolyspora erythraea. ACS Synth. Biol. 2018, 7, 1338–1348.
- Xu, J.; Zhang, J.; Zhuo, J.; Li, Y.; Tian, Y.; Tan, H. Activation and Mechanism of a Cryptic Oviedomycin Gene Cluster Via the Disruption of a Global Regulatory Gene, adpA, in Streptomyces ansochromogenes. J. Biol. Chem. 2017, 292, 19708–19720.
- Thanapipatsiri, A.; Gomez-Escribano, J.P.; Song, L.; Bibb, M.J.; Al-Bassam, M.; Chandra, G.; Thamchaipenet, A.; Challis, G.L.; Bibb, M.J. Discovery of Unusual Biaryl Polyketides by Activation of a Silent Streptomyces venezuelae Biosynthetic Gene Cluster. Chem. Biochem. 2016, 17, 2189–2198.
- Ochi, K. Insights into Microbial Cryptic Gene Activation and Strain Improvement: Principle, Application and Technical Aspects. J. Antibiot. (Tokyo) 2017, 70, 25–40.
- Bu, Q.-T.; Yu, P.; Wang, J.; Li, Z.-Y.; Chen, X.-A.; Mao, X.-M.; Li, Y.-Q.J.M.c.f. Rational Construction of Genome-Reduced and High-Efficient Industrial Streptomyces Chassis Based on Multiple Comparative Genomic Approaches. Microb. Cell. Fact. 2019, 18, 1–17.
- Ikeda, H.; Shin-ya, K.; Omura, S.J. Genome Mining of the Streptomyces avermitilis Genome and Development of Genome-Minimized Hosts for Heterologous Expression of Biosynthetic Gene Clusters. J. Ind. Microbiol. Biotechnol. 2014, 41, 233–250.
- Komatsu, M.; Uchiyama, T.; Ōmura, S.; Cane, D.E.; Ikeda, H. Genome-Minimized Streptomyces Host for the Heterologous Expression of Secondary Metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 2646–2651.
- Myronovskyi, M.; Luzhetskyy, A. Heterologous Production of Small Molecules in the Optimized Streptomyces Hosts. Nat. Prod. Rep. 2019, 36, 1281–1294.
- Myronovskyi, M.; Rosenkränzer, B.; Nadmid, S.; Pujic, P.; Normand, P.; Luzhetskyy, A.J.M.E. Generation of a Cluster-Free Streptomyces albus Chassis Strains for Improved Heterologous Expression of Secondary Metabolite Clusters. Metab. Eng. 2018, 49, 316–324.
- Musiol-Kroll, E.M.; Tocchetti, A.; Sosio, M.; Stegmann, E. Challenges and Advances in Genetic Manipulation of Filamentous Actinomycetes - the Remarkable Producers of Specialized Metabolites. Nat. Prod. Rep. 2019, 36, 1351–1369.
- Loman, N.J.; Pallen, M.J. Twenty Years of Bacterial Genome Sequencing. Nat. Rev. Microbiol. 2015, 13, 787–794.
- Baltz, R.H. Gifted Microbes for Genome Mining and Natural Product Discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588.
- F, D.I.F.; Micheli, G.; Camilloni, G. Restriction Enzymes and Their Use in Molecular Biology: An Overview. J. Biosci. 2019, 44, 38.
- Rittie, L.; Perbal, B. Enzymes Used in Molecular Biology: A Useful Guide. J. Cell Commun. Signal 2008, 2, 25–45.
- Wohlleben, W.; Mast, Y.; Muth, G.; Rottgen, M.; Stegmann, E.; Weber, T. Synthetic Biology of Secondary Metabolite Biosynthesis in Actinomycetes: Engineering Precursor Supply as a Way to Optimize Antibiotic Production. FEBS Lett. 2012, 586, 2171–2176.
- Baltz, R.H. Synthetic Biology, Genome Mining, and Combinatorial Biosynthesis of NRPS-Derived Antibiotics: A Perspective. J. Ind. Microbiol. Biotechnol. 2018, 45, 635–649.
- Aubry, C.; Pernodet, J.-L.; Lautru, S. Modular and Integrative Vectors for Synthetic Biology Applications in Streptomyces Spp. Appl. Environ. Microbiol. 2019, 85, e00485–e00419.
- Li, L.; Liu, X.; Jiang, W.; Lu, Y. Recent Advances in Synthetic Biology Approaches to Optimize Production of Bioactive Natural Products in Actinobacteria. Front. Microbiol. 2019, 10.
- Shetty, R.P.; Endy, D.; Knight, T.F., Jr. Engineering Biobrick Vectors from Biobrick Parts. J. Biol. Eng. 2008, 2, 5.
- Purnick, P.E.; Weiss, R. The Second Wave of Synthetic Biology: From Modules to Systems. Nat. Rev. Mol. Cell Biol. 2009, 10, 410–422.
- Rebets, Y.; Brotz, E.; Tokovenko, B.; Luzhetskyy, A. Actinomycetes Biosynthetic Potential: How to Bridge in Silico and in Vivo? J. Ind. Microbiol. Biotechnol. 2014, 41, 387–402.
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics, 1st ed.; John Innes Foundation: Norwich, England, 2000; pp. 18–581.
