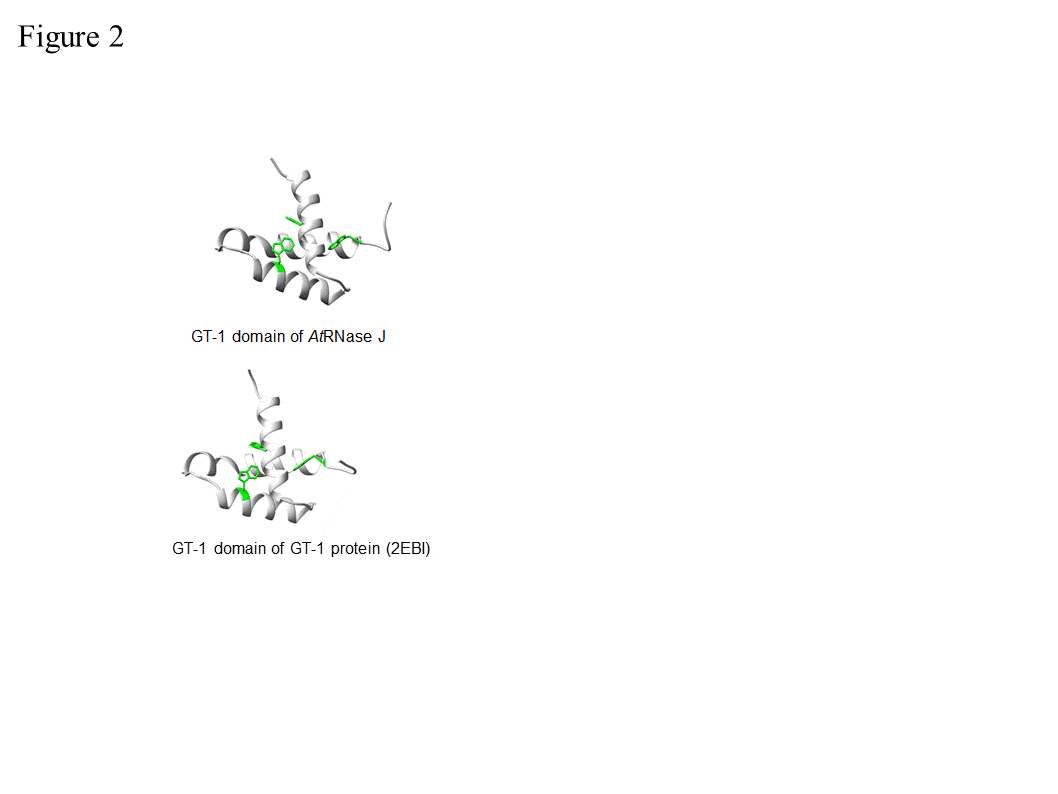RNA quality control is an indispensable but poorly understood process that enables organisms to distinguish functional RNAs from nonfunctional or inhibitory ones. In chloroplasts, whose gene expression activities are required for photosynthesis, retrograde signaling and plant development, RNA quality control is of paramount importance as transcription is relatively unregulated. The functional RNA population is distilled from this initial transcriptome by a combination of RNA-binding proteins and ribonucleases. One of the key enzymes is RNase J, a 5’ - 3’ exoribonuclease, and endoribonuclease, that has been shown to trim 5’ and 3’ RNA termini, and eliminate deleterious antisense RNA. In the absence of RNase J, embryo development cannot be completed. Land plant RNase J contains a highly conserved C-terminal domain that is found in GT-1 DNA-binding transcription factors and is not present in its bacterial, archaeal and algal counterparts. The GT-1 domain may confer specificity through DNA and/or RNA binding and/or protein-protein interactions, and thus be an element in the mechanisms that identify target transcripts among diverse RNA populations. Further understanding of chloroplast RNA quality control relies on discovering how RNase J is regulated, and how its specificity is imparted.
- RNA degradation
- antisense RNA
- PCBF proteins
- RNA processing
This chapter is taken from [1]
1. Ribonuclease J and β-CASP proteins.
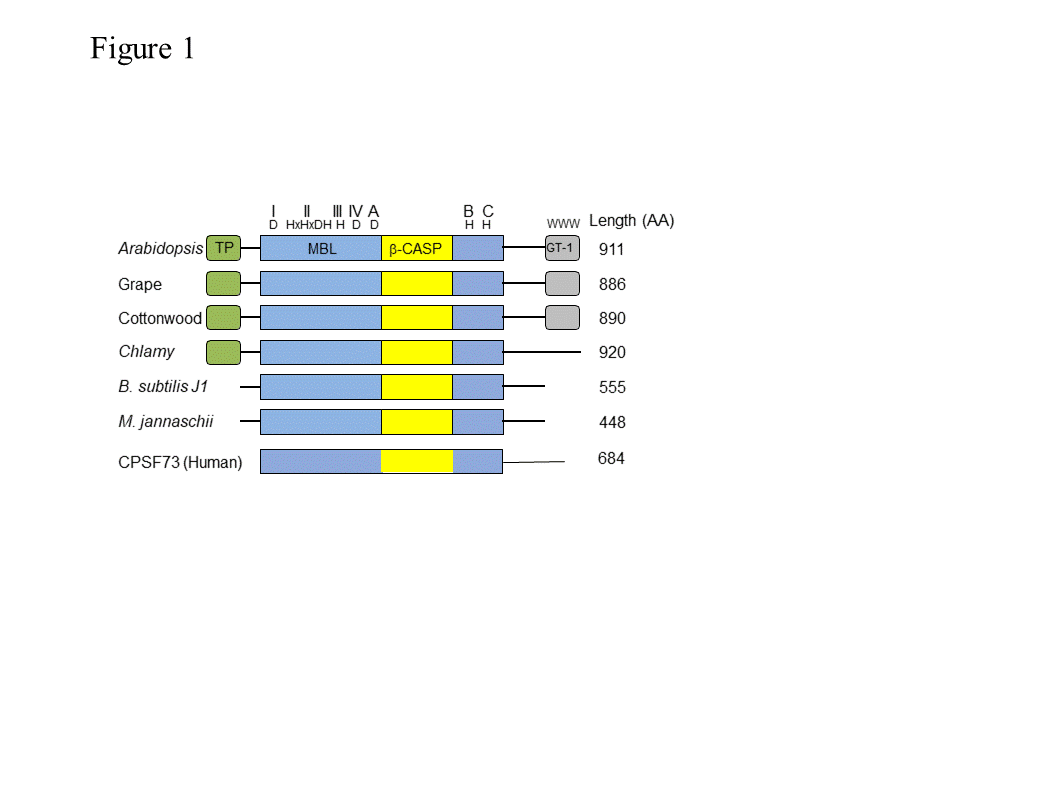
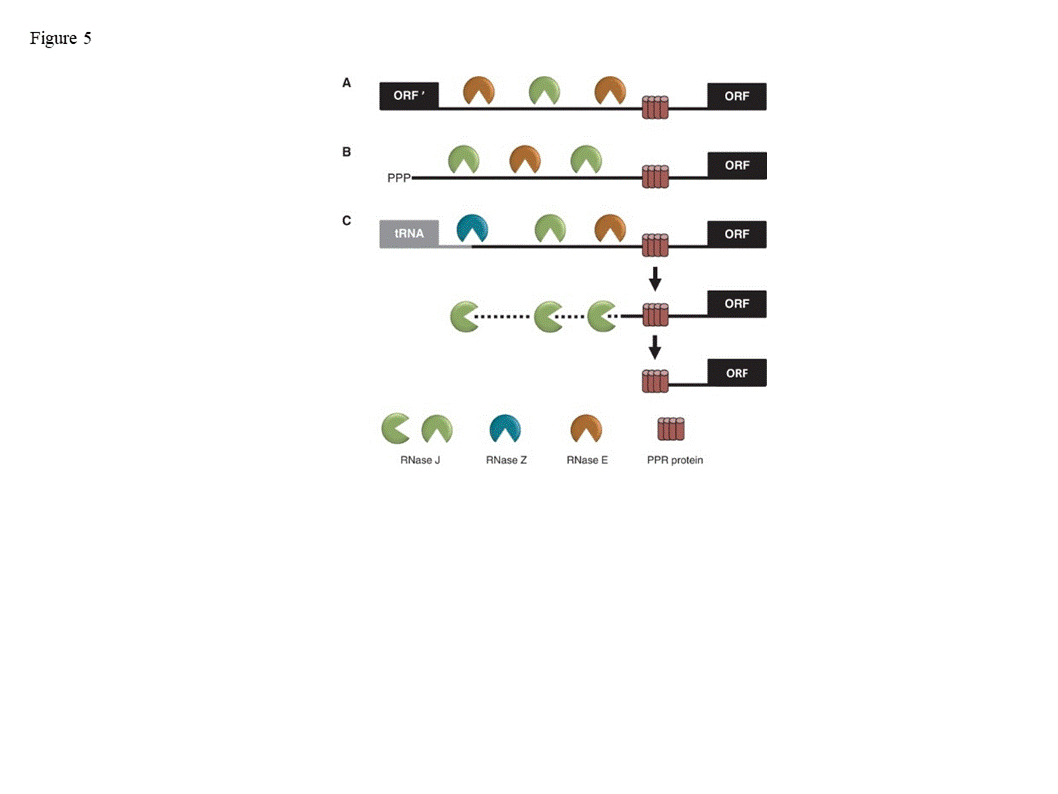
RNase J1 (and the related J2) were first described in B. subtilis [1] [1]. RNase J-CPSF (cleavage and polyadenylation specific factor) homologs are present in most bacteria, Archaea, chloroplasts and eukaryotic cells (Figure 1), suggestive of a ribonuclease that appeared early in evolution [2–5][2][3][4][5]. These proteins belong to a large group denoted “β-CASP”, of which a subgroup of β-CASP ribonucleases harbors dual endo- and 5’®3’ exoribonucleolytic activities. The other β-CASP proteins are involved in DNA repair and recombination as well as other functions [6,7][6][7]. In archaea, the β-CASP ribonuclease subgroup has been further divided into three major groups, two with defined orthologues of the eukaryotic CPSF-73 (see description below) and therefore designated CPSF types, and the other orthologous to bacterial RNase J and therefore designated RNase J type [5,8,9][4][5][8], which includes chloroplast RNase J. The domain structure, length, amino acid sequences, and catalytic mechanism of RNase J and related CPSF proteins are mostly conserved. These proteins contain the seven signature motifs of the metallo-b-lactamase (MBL) and β-CASP domains, I (D), II (HxHxDA), III (H), IV (D), A (D/G), B (H) and C (H) (Figure 1), that together participate in the coordination of two catalytic Zn2+ ions [10,11][9][10]. RNase J is active as a dimer or tetramer, and the amino acid sequence responsible for oligomerization is located at the C-terminus. Plant RNase Js contain, in addition to the MBL-β-CASP motifs, a chloroplast transit peptide at the N-terminus and a conserved GT-1 domain that was previously identified in transcription factors at the C-terminus (discussed below).
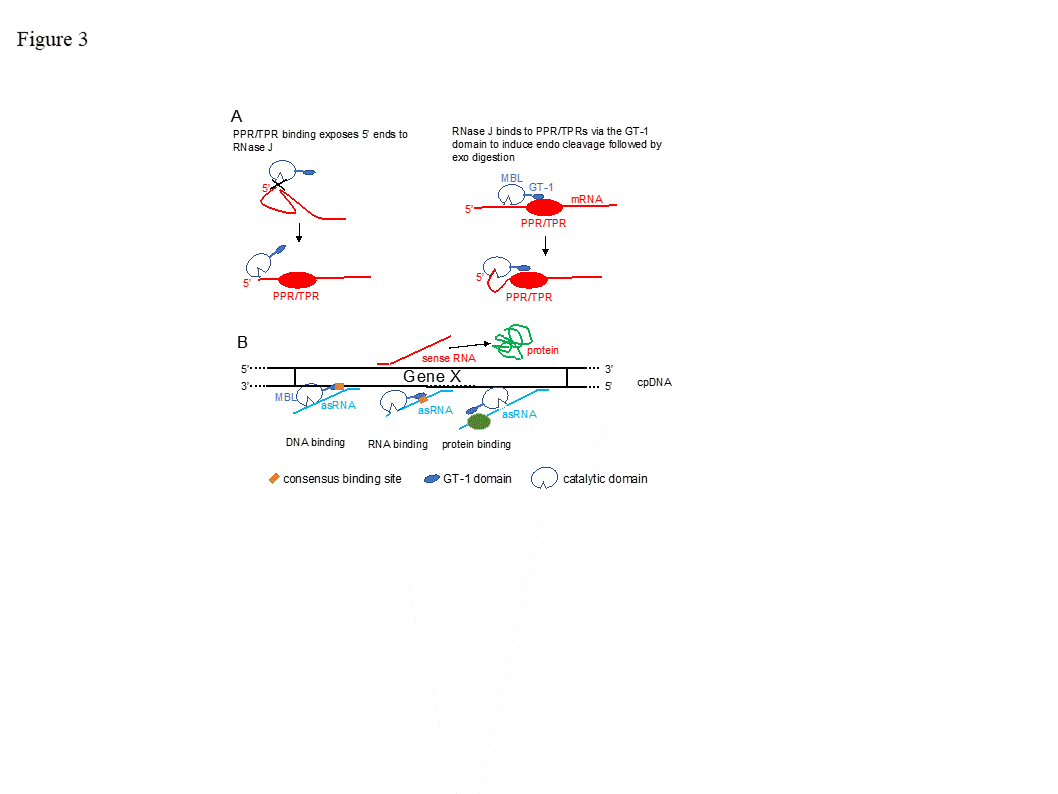
Figure 1. Domain comparison of several plant, bacterial, archaeal and human β-CASP metallo-β-lactamase (MBL) proteins . Arabidopsis RNase J (At5g63420) was used as a query to find homologous proteins. The domain structures from grape (Vitis vinifera ; XM_002279762.1) and cottonwood ( Populus trichocarpa ; XM_002318086.1), representing plants, and from Chlamydomonas reinhardtii (Chlamy), the bacterium Bacillus subtilis (Q45493), and the archaea Methanocaldococcus jannaschii (Q58271) are presented, in comparison with human CPSF-73. The conserved motifs of the metallo-β-lactamase (MBL) and β-CASP (I–IV; A–C) are indicated in blue and yellow, respectively, along with signature amino acid residues (above). Predicted chloroplast transit peptides (TP) are indicated in green. The plant C-terminus includes a region homologous to the GT-1 DNA-binding domain (grey). Its three conserved tryptophan residues are indicated. Crystal structures of bacterial and archaeal RNase J predict a combination of 5’®3’ exonuclease and endonuclease activities, both of which have been observed biochemically in vitro, with the exonuclease activity being dependent on the 5’ end phosphorylation state [10–14][9][10][11][12][13]. Most RNase Js display both 5’®3’ exonucleolytic and endonucleolytic activities when tested in vitro. Chlamydomonas reinhardtii RNase J (CrRNase J) is one of only three family members reported to exhibit exclusively endonucleolytic activity in vitro [2,15,16] [2][14][15]. On the other hand, B. subtilis RNase J1 is mainly exonucleolytic in vitro [17] [16], whereas Arabidopsis RNase J (AtRNase J) displays robust endonucleolytic and relatively minor exonucleolytic activity in vitro [18] [17]. The biological significance and structural basis of the variable exo- and endonucleolytic activities are unknown, but one can predict substrate preferences. For example, exonucleolytic activity might target chemically suitable 5’ RNA termini more efficiently, while an endonuclease could catalyze internal processing or early steps of RNA degradation. In addition to the nature of the RNA 5’ end, the structure of the RNA, as well as other proteins involved, could affect the type of activity carried out by RNase J. The most well studied eukaryotic member of this group is a cleavage and polyadenylation specificity factor of 73 kDa (CPSF-73). This protein is the endonuclease component of a multi-protein complex that plays a key role in pre-mRNA 3'-end formation. It cleaves at a CA motif 20-30 nt downstream of an AAUAAA polyadenylation consensus sequence, and interacts with poly(A) polymerase and other factors to bring about cleavage and polyadenylation of pre-mRNAs in mammalian cells [3,19–21][3][18][19][20]. In addition, it functions as a 5’®3’ exoribonuclease in the maturation of histone pre-mRNA [22][21]. Most archaea encode one or several RNase J/β-CASP homologous proteins and either RNase R or the archaeal exosome. In the group of methanogenic archaea, genes encoding RNase R or the archaeal exosome are not present, suggesting the possibility that RNA processing and degradation is carried out exclusively by RNase J-CPSF proteins [9,15][4][22]. RNase J is present in many, but not all bacteria and those that do not have it, like E. coli, contain the other major endoribonuclease, RNase E. Cyanobacteria that are closely related to the evolutionary ancestor of plant chloroplasts contain both RNase J and RNase E, as do plant chloroplasts, with the exception of the green alga Chlamydomonas reinhardtii, which possesses only RNase J. Endonuclease activity was not identified so far in the degradation of mitochondrial transcripts. However, RNase Z (ELAC2), which is a CPSF homologue, is a mitochondrial endoribonuclease that processes the 3’ end of tRNA precursors. LACTB2 is an endoribonuclease that is present in human mitochondria, belongs to the MBL protein super family and is possibly involved in RNA quality control [23]. RNase P, which is responsible for the 5’ end procession of tRNAs, is an additional mitochondrial endoribonuclease. In spite of their overall conservation with bacterial, archaeal and animal RNase J-CPSF members, plant RNase J’s are distinguished by a C-terminal extension with high homology to the GT-1 DNA-binding domain (Figure 1) [18,24][17][24]. The GT-1 domain was initially defined in pea, and subsequently in ~30-member families of Arabidopsis, wheat and rice transcription factors that regulate various developmental processes and are stress-responsive [25–28][25][26][27][28]. The DNA-binding domain of GT factors features a trihelix structure, each of which contains a conserved tryptophan and amphipathic helix (Figure 2). The GT-1 domain recognizes a degenerate core sequence of 5′-G-Pu-(T/A)-A-A-(T/A)-3′, called the GT element. Such AU-rich sequences are common in intergenic regions of the chloroplast genome. In order to examine the conservation degree of the GT-1 domain in plant RNase J, its predicted structure was superimposed on the known GT-1 transcription factor PDB 2EBI (Figure 2). The DNA-GT-1 interface was located exactly as predicted by the conserved, electropositive, tryptophan-rich interface [18,29]. The predicted structure also displayed similar physicochemical characteristics and a conserved DNA binding site. GT-1-containing transcription factors bind specific nuclear promoter sequences [28], making its presence in plant RNase J somewhat surprising. However, the structural conservation and retention of key residues hint that the GT-1 domain is functional in the context of RNase J. The function of the GT-1 domain in plant RNase J remains enigmatic. While deletion of the GT-1 domain did not interfere with degradation activity in vitro when a purified recombinant protein was incubated with synthetic RNAs [18], it’s more likely in vivo function would be related to sequence specificity, interaction with a PPR protein and/or dimerization, which have not yet been rigorously tested. These possibilities are illustrated in Figure 3. For example, in mRNA 5’ end processing, the GT-1 domain could direct RNase J to certain locations on the RNA by direct sequence-specific binding or by binding to a sequence-specific cofactor (Figure 3 panel A). In the process of removing antisense transcripts, the GT-1 domain could influence RNase J target preference through its DNA, RNA or protein binding properties (Figure 3 panel B and see below). Figure 2. Comparative GT-1 domain structures. Top, three helix model of amino acids (815-874) of the Arabidopsis RNase J (AtRNase J) GT-1 domain, based on (bottom) PDB 2EBI (GT-1 transcription factor amino acids 81-152), built using the NMR-solved structure as a template [29]. The three conserved tryptophans are in green. (Acquired with copyright permission from [18][17]). Figure 3. Models for RNase J modes of action. (A) Two scenarios for chloroplast 5’ end maturation by RNase J and the corresponding RNA-binding protein (RBP). Left , the RNA 5’ end structure prevents RNase J access. Binding of the RBP induces structural change exposing the 5’ end to digestion. Right , RNase J is recruited to the 5’ end by direct binding to the RBP, perhaps via the GT-1 domain. ( B) Possible mechanisms of GT-1 domain-mediated recruitment of RNase J to targeted asRNAs by binding to a DNA site near the asRNA transcription start (left), to the asRNA itself (center), or to an RNA-binding cofactor (right). Possible mechanisms of GT-1 domain-mediated recruitment of RNase J to targeted asRNAs by binding to a DNA site near the asRNA transcription start (left), to the asRNA itself (center), or to an RNA-binding cofactor (right). The only photosynthetic organisms in which an RNase J mutant phenotype has been studied are tobacco and Arabidopsis. Arabidopsis null mutants for RNase J are embryo-lethal, displaying albino ovules containing aborted embryos [30]. Further examination suggested that RNase J is required for the organization and functioning of the shoot apical meristems, cotyledons and hypocotyls [31]. In addition, the transport and response of auxin was impaired [31]. Why absence of RNase J activity results in embryo lethality is still obscure, however the importance of plastid gene expression for embryo maturation in plants is well documented [32,33][32][33]. It is possible that simply impaired functioning of the chloroplast in general (see below), a specific function in the procession or degradation of a particular transcript, or another function that is not related to the ribonuclease activity is responsible for embryo lethality. In general, AtRNase J is highly expressed in cells containing chloroplasts, as well as in reproductive organs, and its expression is significantly light-dependent [31]. Because RNase J null mutants are embryo-lethal, virus-induced gene silencing (VIGS) was used to decrease RNase J abundance in tobacco and Arabidopsis [24,34][24][34]. The most striking effect of RNase J deficiency was massive accumulation of asRNAs, suggesting that the previously-documented failure of chloroplast RNA polymerase to terminate efficiently [35] [35] leads to symmetric transcription products that are normally eliminated by RNase J (Figure 4). This situation is exacerbated because chloroplast genomes are compact, with what generally appears to be a random distribution of genes on one strand versus another. In RNase J-down-expressed tissues, antisense-sense duplexes were readily detected, and correlated with failure to associate with polysomes, chlorosis and tissue death [24]. Therefore, in addition to its function in 5’ end processing, RNase J appears to play a major and essential role in chloroplast RNA quality control by eliminating long and otherwise abundant antisense transcripts. Open questions remain, however, as to whether rapid elimination of antisense transcripts is required for the successful translation of the sense strand transcripts. It has long been known that transcription termination at the 3’ end of most genes is inefficient in chloroplasts, necessitating RNA maturation mechanisms to create defined 3’ termini [35]. Since chloroplast genomes are compact, and in most cases have an apparently random distribution of genes on one strand versus another, potential accumulation of double stranded molecules formed by sense and antisense transcripts is high. This situation is harmful for translation, therefore the antisense transcript would normally be rapidly eliminated [24]. The plant chloroplast RNase J has assumed the role of RNA surveillance, eliminating the antisense transcripts (Figure 4). Whether the GT-1 domain is important in this function, and more globally how RNase J differentiates between sense and antisense RNA to rapidly remove the second is still obscured. A possible scenario imposing the plant specific GT-1 domain in thisprocess is presented in Figures 3. Figure 4. Model for antisense RNA (asRNA) surveillance by chloroplast RNase J. (A) 3′ UTRs of chloroplast genes inefficiently terminate transcription, resulting in read-through (mRNA-1 and mRNA-2). Where genes are convergently transcribed, even at a distance, asRNA may be synthesized. (B) These pre-mRNAs are first processed by an endonuclease, which could possibly be RNase J itself, or another unidentified endonuclease. This creates substrates for the 5′ → 3′ exonucleolytic activity of RNase J. (C) By removing asRNA, RNase J allows accumulation of single-stranded sense RNA that is translationally competent mRNAs. (Acquired with copyright permission from [24]) Analysis of VIGS-induced RNase J knockdown in plant tissue revealed that, in addition to its role in eliminating antisense transcripts, RNase J matures the 5’ ends of several transcripts, being guided or blocked by PPR proteins, as previously postulated [34,36] [34][36] and consistent with in vitro analysis of its catalytic activity [18,24] [17][24]. The 5’ maturation could occur by RNase J-catalyzed endonucleolytic cleavage followed by 5’®3’ exonuclease degradation until blocked by the corresponding PPR protein, generating the mature transcript 5’ end (Figure 5) [36]. The observed robust endoribonucleolytic and exonucleolytic activity of RNase J in vitro using purified recombinant enzyme and a synthetic RNA, supports that possibility. Otherwise, if RNase J is active in vivo exclusively as an exonuclease, the endonucleolytic cleavage could be performed by another endoribonuclease such as RNase E or RNase Z, followed by 5’®3’ exoribonucleolytic processive degradation by RNase J. Figure 5 illustrates the various modes of RNase J participation in chloroplast RNA 5’ end maturation, depending on whether the substrates are derived from intercistronic cleavage, nearby transcription initiation, or 3’ processing of an upstream tRNA. Figure 5. Model for the involvement of RNase J in processing chloroplast RN Model for the involvement of RNase J in processing chloroplast RNA 5’ ends defined by PPR proteins. Maturation of a generic mRNA (ORF) 5’ end is shown here. Precursor transcripts originate from polycistronic ( A 5’ ends defined by PPR proteins. Maturation of a generic mRNA (ORF) 5’ end is shown here. Precursor transcripts originate from polycistronic (A) and monocistronic (B) transcriptional units, as well as readthrough transcripts from upstream genes such as tRNAs (C). Processing is initiated by endonucleolytic cleavages by RNase J or RNase E within unstructured intergenic regions, or in the case of tRNAs by RNase Z. The resultant 5’ ends are subsequently trimmed to their mature forms by the exonuclease activity of RNase J to PPR protein-bound sites. (Acquired with copyright permission from [34]). ) and monocistronic (B) transcriptional units, as well as readthrough transcripts from upstream genes such as tRNAs (C). Processing is initiated by endonucleolytic cleavages by RNase J or RNase E within unstructured intergenic regions, or in the case of tRNAs by RNase Z. The resultant 5’ ends are subsequently trimmed to their mature forms by the exonuclease activity of RNase J to PPR protein-bound sites. (Acquired with copyright permission from [34]).2. The plant RNase J GT-1 domain
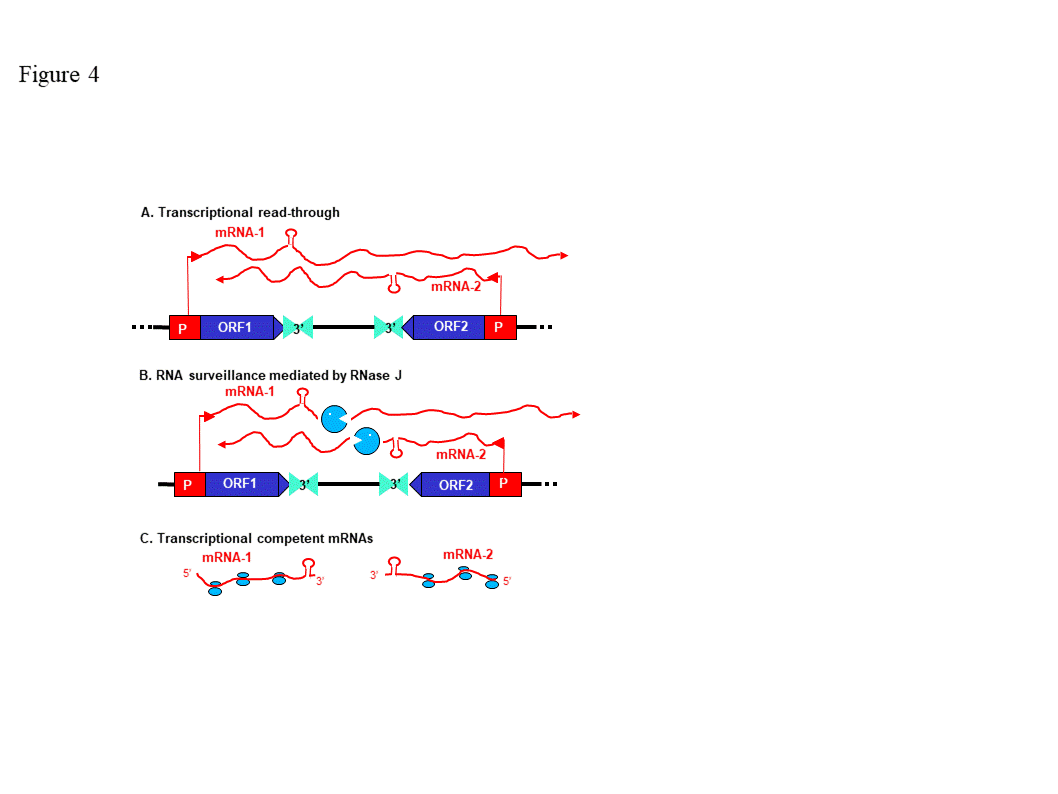
3. Consequences of removing or down-regulating RNase J in plants
References
- Amber M. Hotto; David B. Stern; Gadi Schuster; Plant Ribonuclease J: An Essential Player in Maintaining Chloroplast RNA Quality Control for Gene Expression. PSergine Even; Olivier Pellegrini; Lena Zig; Valerie Labas; Joelle Vinh; Dominique Bréchemmier-Baey; Harald Putzer; Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Reseants rch 2020, 9, 334, 10.3390/plants9030334.5, 33, 2141-2152, 10.1093/nar/gki505.
- Ciarán Condon; Laetitia Gilet; The Metallo-β-Lactamase Family of Ribonucleases. Recoding: Expansion of Decoding Rules Enriches Gene Expression 2011, null, 245-267, 10.1007/978-3-642-21078-5_10.
- Dominski, Z.; Carpousis, A.J.; Clouet-d’Orval, B. Emergence of the beta-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta 2013, 1829, 532–551.
- Béatrice Clouet-D’Orval; Manon Batista; Marie Bouvier; Yves Quentin; Gwennaële Fichant; Anita Marchfelder; Lisa-Katharina Maier; Insights into RNA-processing pathways and associated RNA-degrading enzymes in Archaea. FEMS Microbiology Reviews 2018, 42, 579-613, 10.1093/femsre/fuy016.
- Duy Khanh Phung; Dana Rinaldi; Petra Langendijk-Genevaux; Yves Quentin; Agamemnon Carpousis; Béatrice Clouet-D’Orval; Archaeal β-CASP ribonucleases of the aCPSF1 family are orthologs of the eukaryal CPSF-73 factor.. Nucleic Acids Research 2012, 41, 1091-103, 10.1093/nar/gks1237.
- I. Callebaut; Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Research 2002, 30, 3592-3601, 10.1093/nar/gkf470.
- Zbigniew Dominski; Nucleases of the Metallo-β-lactamase Family and Their Role in DNA and RNA Metabolism. Critical Reviews in Biochemistry and Molecular Biology 2007, 42, 67-93, 10.1080/10409230701279118.
- Béatrice Clouet-D’Orval; Duy Khanh Phung; Petra Langendijk-Genevaux; Yves Quentin; Universal RNA-degrading enzymes in Archaea: Prevalence, activities and functions of β-CASP ribonucleases. Biochimie 2015, 118, 278-285, 10.1016/j.biochi.2015.05.021.
- Xin Zheng; Na Feng; Defeng Li; Xiuzhu Dong; Jie Li; New molecular insights into an archaeal RNase J reveal a conserved processive exoribonucleolysis mechanism of the RNase J family. Molecular Microbiology 2017, 106, 351-366, 10.1111/mmi.13769.
- Xue-Yuan Pei; Patricia Bralley; G H Jones; Ben F. Luisi; Linkage of catalysis and 5' end recognition in ribonuclease RNase J.. Nucleic Acids Research 2015, 43, 8066-76, 10.1093/nar/gkv732.
- Inès Li De La Sierra-Gallay; Léna Zig; Ailar Jamalli; Harald Putzer; Structural insights into the dual activity of RNase J. Nature Structural & Molecular Biology 2008, 15, 206-212, 10.1038/nsmb.1376.
- Audrey Dorléans; Inès Li De La Sierra-Gallay; Jérémie Piton; Léna Zig; Laetitia Gilet; Harald Putzer; Ciarán Condon; Molecular Basis for the Recognition and Cleavage of RNA by the Bifunctional 5′–3′ Exo/Endoribonuclease RNase J. Structure 2011, 19, 1252-1261, 10.1016/j.str.2011.06.018.
- Condon, C. What is the role of RNase J in mRNA turnover? RNA Biol. 2010, 7, 316–321.
- Levy, S.; Portnoy, V.; Admon, J.; Schuster, G. Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biol. 2011, 8, 1073–1083.
- Liponska, A.; Jamalli, A.; Kuras, R.; Suay, L.; Garbe, E.; Wollman, F.-A.; Laalami, S.; Putzer, H. Tracking the elusive 5 ′ exonuclease activity of Chlamydomonas reinhardtii RNase J. Plant Mol. Biol. 2018, 96, 641–653.
- Nathalie Mathy; Lionel Benard; Olivier Pellegrini; Roula Daou; Tingyi Wen; Ciarán Condon; 5′-to-3′ Exoribonuclease Activity in Bacteria: Role of RNase J1 in rRNA Maturation and 5′ Stability of mRNA. Cell 2007, 129, 681-692, 10.1016/j.cell.2007.02.051.
- Michal Halpert; Varda Liveanu; Fabian Glaser; Gadi Schuster; The Arabidopsis chloroplast RNase J displays both exo- and robust endonucleolytic activities. Plant Molecular Biology 2018, 99, 17-29, 10.1007/s11103-018-0799-5.
- Corey R. Mandel; Syuzo Kaneko; Hailong Zhang; Damara Gebauer; Vasupradha Vethantham; James L Manley; Liang Tong; Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease. Nature 2006, 444, 953-956, 10.1038/nature05363.
- L. L. Koekemoer; Maureen Coetzee; R. H. Hunt; Hpall endonuclease distinguishes between two species in the Anopheles funestus group.. Insect Molecular Biology 1998, 7, 273-277, 10.1046/j.1365-2583.1998.00072.x.
- Serena Chan; Eun-A Choi; Yongsheng Shi; Pre-mRNA 3'-end processing complex assembly and function.. Wiley Interdisciplinary Reviews: RNA 2010, 2, 321-35, 10.1002/wrna.54.
- Xiao-Cui Yang; Kelly D Sullivan; William F. Marzluff; Zbigniew Dominski; Studies of the 5′ Exonuclease and Endonuclease Activities of CPSF-73 in Histone Pre-mRNA Processing. Molecular and Cellular Biology 2008, 29, 31-42, 10.1128/mcb.00776-08.
- Shiri Levy; Victoria Portnoy; Jasmine Admon; Gadi Schuster; Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biology 2011, 8, 1073-1083, 10.4161/rna.8.6.16604.
- Levy, S.; Allerston, C.K.; Liveanu, V.; Habib, M.R.; Gileadi, O.; Schuster, G. Identification of LACTB2, a metallo-beta-lactamase protein, as a human mitochondrial endoribonuclease. Nucleic Acids Res. 2016, 44, 1813–1832.
- Robert Sharwood; Michal Halpert; Scott Luro; Gadi Schuster; David B. Stern; Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA 2011, 17, 2165-2176, 10.1261/rna.028043.111.
- Zhanchao Wang; Quangang Liu; Hanzeng Wang; Haizhen Zhang; Xuemei Xu; Chenghao Li; Chuanping Yang; Comprehensive analysis of trihelix genes and their expression under biotic and abiotic stresses in Populus trichocarpa. Scientific Reports 2016, 6, 36274, 10.1038/srep36274.
- Wenli Wang; Peng Wu; TongKong Liu; Haibo Ren; Ying Li; Xilin Hou; Genome-wide Analysis and Expression Divergence of the Trihelix family in Brassica Rapa: Insight into the Evolutionary Patterns in Plants. Scientific Reports 2017, 7, 6463, 10.1038/s41598-017-06935-0.
- Jie Xiao; Rui Hu; Ting Gu; Jiapeng Han; Ding Qiu; Peipei Su; Jialu Feng; Junli Chang; Guangxiao Yang; Guangyuan He; Genome-wide identification and expression profiling of trihelix gene family under abiotic stresses in wheat. BMC Genomics 2019, 20, 287, 10.1186/s12864-019-5632-2.
- Ruth N. Kaplan-Levy; Philip Brewer; Tezz Quon; David Smyth; The trihelix family of transcription factors – light, stress and development. Trends in Plant Science 2012, 17, 163-171, 10.1016/j.tplants.2011.12.002.
- Takashi Nagata; Emi Niyada; Natsuki Fujimoto; Yuuya Nagasaki; Kazuaki Noto; Youhei Miyanoiri; Jun Murata; Kazuyuki Hiratsuka; Masato Katahira; Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: Mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins: Structure, Function, and Bioinformatics 2010, 78, 3033-3047, 10.1002/prot.22827.
- Iris Tzafrir; Rosanna Pena-Muralla; Allan Dickerman; Michael Berg; Rebecca Rogers; Steven Hutchens; T. Colleen Sweeney; John McElver; George Aux; David Patton; David W. Meinke; Identification of Genes Required for Embryo Development in Arabidopsis1[w]. Plant Physiology 2004, 135, 1206-1220, 10.1104/pp.104.045179.
- Hongyu Chen; Wenxuan Zou; Jie Zhao; Ribonuclease J is required for chloroplast and embryo development in Arabidopsis.. Journal of Experimental Botany 2015, 66, 2079-91, 10.1093/jxb/erv010.
- Ningning Yuan; Jiechen Wang; Yong Zhou; Dong An; Qiao Xiao; Wenqin Wang; Yongrui Wu; EMB-7L is required for embryogenesis and plant development in maize involved in RNA splicing of multiple chloroplast genes.. Plant Science 2019, 287, 110203, 10.1016/j.plantsci.2019.110203.
- Shih-Chi Hsu; Mark F Belmonte; John J Harada; Kentaro Inoue; Indispensable Roles of Plastids in Arabidopsis thaliana Embryogenesis. Current Genomics 2010, 11, 338-349, 10.2174/138920210791616716.
- Scott Luro; Arnaud Germain; Robert Sharwood; David B. Stern; RNase J participates in a pentatricopeptide repeat protein-mediated 5' end maturation of chloroplast mRNAs.. Nucleic Acids Research 2013, 41, 9141-51, 10.1093/nar/gkt640.
- Arnaud Germain; Amber M. Hotto; Alice Barkan; David B. Stern; RNA processing and decay in plastids. Wiley Interdisciplinary Reviews: RNA 2013, 4, 295-316, 10.1002/wrna.1161.
- Jeannette Pfalz; Omer Ali Bayraktar; Jana Prikryl; Alice Barkan; Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. The EMBO Journal 2009, 28, 2042-2052, 10.1038/emboj.2009.121.