Chronic psychological stress is known to have physiological impacts and may involve epigenetic modifications. We investigated whether individuals with consistently high self-reported stress levels exhibit differential DNA methylation at specific CpG sites that have been associated with fibromyalgia, a chronic pain syndrome. Saliva DNA from two cohorts – a high-stress group (N = 357, 63% male, mean age 42 ± 1.8) and a low-stress comparison group (N = 37, 58% male, mean age 41 ± 2.0) – was analyzed using an Illumina Infinium methylation microarray (EPIC v2 “Mercury” beadchip) covering >850,000 CpG sites. We focused on three specific CpG loci (cg15957394, cg13348458, cg25294185) previously identified as significantly hypomethylated in fibromyalgia patients compared to healthy controls. The average methylation across these sites (a composite “Epi Vitality” Muhdo Health index) was compared between groups. The high-stress cohort showed a significantly lower mean methylation (mean β = 0.248) than the low-stress group (mean β = 0.271, Mann–Whitney U test p = 0.0225). Median methylation in the high-stress group was 0.233 (interquartile range, IQR, 0.195–0.291) vs. 0.280 (IQR 0.217–0.310) in low-stress subjects. This indicates that chronic high stress is associated with a subtle but significant DNA hypomethylation signature in saliva, similar in direction to that observed in fibromyalgia. Our findings provide preliminary evidence linking psychological stress with specific epigenetic changes that have been implicated in chronic pain, supporting the hypothesis that chronic stress and fibromyalgia may share common epigenetic pathways.
- Stress
- Psychology
- epigenetics
- Emotional Stress
- fibromyalgia
- pain
- CFS
- fatigue
- DNA methylation
1. Introduction
Chronic stress has pervasive effects on human biology and has been associated with lasting epigenetic changes. In fact, many studies have shown that prolonged stress exposures can leave “imprints” on the epigenome, particularly in genes related to the stress response [1]. For example, genes such as SLC6A4 (serotonin transporter) and BDNF (brain-derived neurotrophic factor) have been reported to undergo stress-induced DNA methylation alterations in individuals experiencing high stress. These stress-related epigenetic modifications may contribute to downstream physiological and psychological effects, potentially influencing an individual’s risk for stress-related disorders.
Fibromyalgia (FM) is a chronic pain condition that has also been linked to aberrant epigenetic patterns. Emerging evidence indicates that fibromyalgia patients exhibit DNA hypomethylation at specific loci compared to healthy controls [2]. In one epigenome-wide study of saliva DNA, five autosomal CpG sites were identified as significantly less methylated in fibromyalgia patients, relative to non-FM controls. Notably, these top fibromyalgia-associated CpGs map to genes involved in cytoskeletal signalling (AFAP1), immune DNA sensing (RNASEH2C), a ubiquitin-pathway pseudogene (UBE2Q2P1), and two intergenic regions. Such genes and pathways align with processes of neuro-immune modulation and stress response, suggesting a possible overlap between fibromyalgia’s epigenetic signature and the biological effects of chronic stress. Fibromyalgia’s hypomethylation profile is enriched in genes related to stress response and DNA repair pathways [3], hinting that chronic stress might be one factor contributing to these epigenetic changes.
Given this background, we hypothesized that individuals experiencing high levels of daily stress would display DNA methylation differences similar to those seen in fibromyalgia. In particular, we focused on a set of three CpG sites – cg15957394, cg13348458, and cg25294185 – which are part of an epigenetic biomarker panel (the “Epi Vitality” test by Muhdo Ltd.) derived from the aforementioned fibromyalgia study. Hypomethylation at these three loci (located in or near AFAP1, an intergenic region on chromosome 6, and RNASEH2C, respectively) has been linked to fibromyalgia and increased pain susceptibility. We aimed to determine whether a similar hypomethylation pattern could be observed in people under chronic high stress, compared to those with minimal stress, thereby exploring a potential epigenetic bridge between psychological stress and chronic pain conditions.
2. Materials and Methods
Study Cohorts: Two adult cohorts were recruited based on self-reported daily stress levels. The Focus (High Stress) group consisted of 357 individuals who consistently rated their perceived stress as “very high” every day. The Complement (Low Stress) group included 37 individuals who reported “very low” or no daily stress. Participants in both groups were of similar age (mean ages ~42 and ~41 years, respectively) and had a comparable sex distribution (≈60–63% male in each group). All participants provided informed consent for saliva collection and genetic/epigenetic analysis. To avoid confounding factors, individuals with any diagnosed chronic illness (aside from stress or stress-related symptoms) were excluded, so that neither cohort included fibromyalgia patients or other known pathological conditions.
Saliva DNA Methylation Profiling: Unstimulated saliva samples were collected from each participant using standard DNA collection kits. Genomic DNA was extracted from saliva and for DNA methylation analysis. We employed the Illumina Infinium MethylationEPIC array. Array processing followed manufacturer protocols and established quality control pipelines. Raw methylation beta values (β, representing the fraction of DNA molecules methylated at a given CpG site) were obtained for each probe.
Targeted 3-CpG Panel and Composite Score: Rather than conducting an epigenome-wide association test, we focused on three specific CpG sites of interest (cg15957394, cg13348458, cg25294185). These loci were selected based on prior findings of significant hypomethylation in fibromyalgia patients, and together they form an epigenetic health panel known as “Epi Vitality.” Table 1 summarizes the genomic context of these CpGs. For each participant, we extracted the beta values at these three probes and calculated their mean as a composite methylation index. This 3-CpG average represents the individual’s overall methylation level across the panel and served as our primary outcome measure. By condensing the three loci into one metric, we aimed to improve signal-to-noise ratio and examine group differences in a holistic manner (analogous to the 5-CpG mean metric used in the fibromyalgia study.)
Table 1. Targeted CpG sites used in this study (“EpiVitality” 3-CpG panel), with their genomic annotations.
|
CpG ID |
Gene Annotation |
Genomic Location |
Region |
Reported in Fibromyalgia |
|
cg15957394 |
AFAP1 (Actin Filament-Associated Protein 1) – promoter region |
chr4:7940563-7941853 |
Promoter |
Yes – 7.2% lower methylation in FM |
|
cg13348458 |
Intergenic (no nearby gene) |
chr6:27569325-27570075 |
Intergenic |
Yes – hypomethylated in FM |
|
cg25294185 |
RNASEH2C (RNase H2, subunit C) – promoter |
chr11:65487340-65487875 |
Promoter |
Yes – 3.5% lower methylation in FM |
Note: FM = fibromyalgia. Prior research identified these three loci (as part of a five-CpG panel) as significantly hypomethylated in fibromyalgia patients relative to controls. Lower methylation at these sites has been linked to fibromyalgia’s pathophysiology and pain levels.
Statistical Analysis: Group differences in the 3-CpG composite methylation index were assessed using the non-parametric Mann–Whitney U test (two-tailed). This test was chosen because methylation beta values are often not normally distributed and the group sizes were highly unequal (n_high=357 vs n_low=37). The null hypothesis was that the distribution of the 3-CpG average methylation is the same in both high-stress and low-stress groups. A p-value < 0.05 was considered statistically significant. Given that the three CpGs were selected based on prior evidence, we treated this as a targeted analysis; however, we also report the false discovery rate (FDR) q-value for the group comparison. The FDR was calculated via the Benjamini–Hochberg method, though with only one primary comparison (the composite index) the q-value is essentially equal to the raw p-value (q = 0.0225). Additionally, we examined basic distribution statistics (median, interquartile range, minima/maxima) for the methylation index in each group.
3. Results
Group Methylation Differences: A significant difference in DNA methylation was observed between individuals with high daily stress and those with low stress. The average methylation across the three CpG sites (Epi Vitality index) was lower in the high-stress Focus cohort (mean β = 0.248) compared to the low-stress Complement cohort (mean β = 0.271). This corresponds to an absolute difference of about –0.023 (–2.34 percentage points) in the mean beta value between the groups. Despite the modest magnitude, this difference was statistically significant (Mann–Whitney U p = 0.0225; FDR-adjusted q = 0.0225). The null hypothesis of equal distributions was thus rejected, indicating that the high-stress group tends to have lower methylation at these loci than the low-stress group.
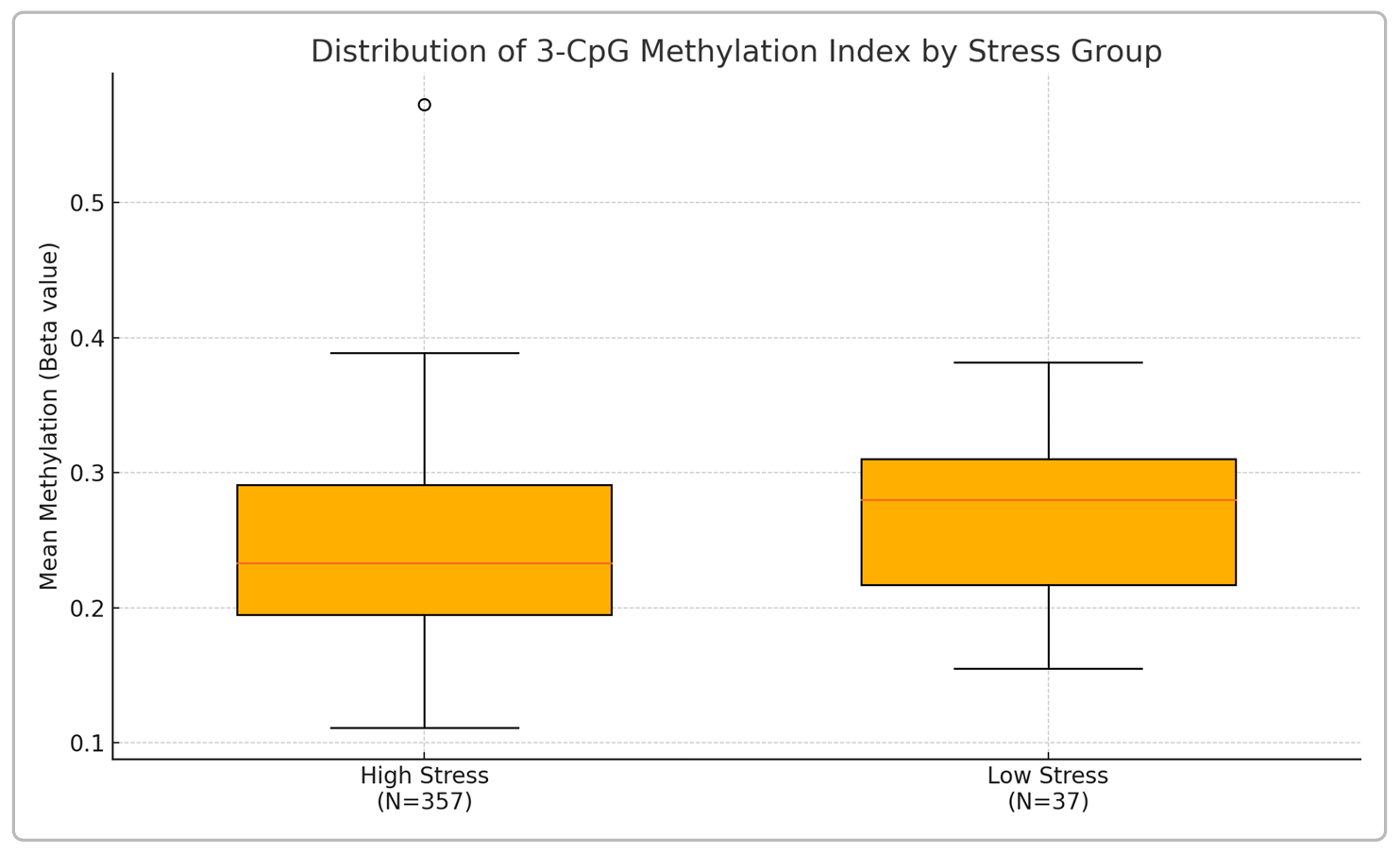
Figure 1. Box plot showing the distribution of the 3-CpG composite methylation index in the high-stress (Focus) and low-stress (Complement) groups. Each box spans the 25th–75th percentile (IQR) of the methylation values, with the median indicated by the red line. Whiskers extend to the 5th and 95th percentiles; points beyond these (e.g., the open circle above the high-stress group) represent outliers. The high-stress group exhibits a notably lower median and overall shift toward lower DNA methylation compared to the low-stress group. This visual summary illustrates the group difference in methylation levels associated with chronic stress.
Table 2 provides summary statistics of the methylation index in each cohort. The high-stress group had a median composite methylation of 0.233 (with an interquartile range, IQR, of 0.195–0.291). In contrast, the low-stress group had a higher median of 0.280 (IQR 0.217–0.310). The entire distribution for high-stress individuals was left-shifted toward lower methylation values relative to low-stress individuals, as also depicted in Figure 1. Notably, the spread of methylation values was somewhat wider in the high-stress group (range 0.111 to 0.573) than in the low-stress group (range 0.155 to 0.394). In the high-stress cohort, a small number of individuals had very low composite methylation (down to ~0.11) or relatively high values (~0.50–0.57) representing outliers, whereas the low-stress cohort’s values were more tightly clustered (most between 0.20 and 0.38). The medians and IQRs, however, indicate a clear central tendency difference, with the high-stress median ~4.7 percentage points lower than the low-stress median. This median gap is of similar order to what has been reported between fibromyalgia patients and controls using a larger 5-CpG panel (median difference ~6.1 percentage points in one study), though our high-stress vs low-stress difference is smaller in magnitude.
Table 2. Composite DNA methylation index (mean β value of 3 CpGs) in high-stress vs low-stress groups.
|
Group |
N |
Mean β |
Median β (IQR) |
Min – Max |
|
High Stress (Focus) |
357 |
0.248 |
0.233 (0.195 – 0.291) |
0.111 – 0.573 |
|
Low Stress (Complement) |
37 |
0.271 |
0.280 (0.217 – 0.310) |
0.155 – 0.394 |
A non-parametric comparison confirmed that the distribution of methylation values differs significantly between the two groups (U test p = 0.0225). In practical terms, individuals reporting very high daily stress tend to have a lower epigenetic “score” at these three fibromyalgia-linked CpG sites, whereas those with minimal stress have higher methylation on average. We also examined each CpG site individually: in all three cases, the high-stress group showed lower median β values than the low-stress group (data not shown). However, because the three CpGs are correlated in their methylation pattern (as they likely respond to similar physiological influences), the composite index provides a more robust measure of the overall difference. No other covariates (such as age or sex) showed any significant association with the 3-CpG methylation levels in this dataset (age was narrowly distributed around ~42 in both groups, and sex effects were minimized by focusing on autosomal loci).
4. Discussion
In this study, we found that people experiencing chronic high stress have significantly lower DNA methylation at a set of CpG sites that have been previously implicated in fibromyalgia. This result supports our hypothesis that chronic stress is linked to DNA hypomethylation at specific genomic loci, mirroring patterns seen in a chronic pain disorder. The three CpG sites we examined were originally discovered as part of a fibromyalgia-associated epigenetic signature. Our findings extend this concept by showing that even in individuals without fibromyalgia, those who report high psychological stress show an epigenetic shift in the same direction (hypomethylation) at these sites, compared to low-stress individuals. This suggests a possible shared epigenetic pathway between chronic stress and fibromyalgia/pain conditions.
The magnitude of the methylation difference we observed (~2–4% lower in high stress vs low stress, depending on measure) is smaller than the difference reported between full-fledged fibromyalgia patients and controls (~5–7% lower in FM). This is perhaps not surprising: fibromyalgia is a severe clinical syndrome, and the epigenetic changes in patients might be more pronounced due to the intensity and chronicity of pain, potentially compounding stress-related effects. In contrast, our high-stress participants, while significantly stressed, may not all have developed somatic conditions like fibromyalgia. The fact that we still detect a clear hypomethylation trend in the high-stress group points to stress alone being sufficient to drive detectable epigenetic change, albeit of a moderate degree. It raises the question of causality and progression – for instance, could prolonged extreme stress lead to greater methylation aberrations over time, eventually reaching the levels observed in fibromyalgia? Longitudinal studies would be needed to explore whether continued stress worsens this epigenetic divergence and whether individuals with more extreme hypomethylation are at higher risk of developing stress-related disorders (including chronic pain syndromes).
Mechanistically, the loci involved provide clues that align with known stress physiology and fibromyalgia pathophysiology. AFAP1 (Actin Filament-Associated Protein 1), associated with cg15957394, is an adaptor protein in cytoskeletal signalling pathways and has been identified as a key mediator in certain inflammatory signalling processes. Hypomethylation in a gene’s promoter often correlates with increased gene expression; thus, lower methylation at the AFAP1 promoter in high-stress individuals might imply upregulation of AFAP1. If AFAP1 is indeed upregulated, it could modulate immune cell behaviour or inflammatory signal transduction, since AFAP1 has been linked to TNF-α signalling in endothelial cells. Chronic stress is known to provoke low-grade inflammation and immune dysregulation (e.g., via elevated cortisol, catecholamines, and sympathetic nervous system activation). An epigenetic upregulation of AFAP1 could therefore be part of the molecular interface between psychological stress and the immune/inflammatory system – a connection that is also thought to be relevant in fibromyalgia’s pathology.
The second gene highlighted is RNASEH2C, corresponding to cg25294185 in its promoter. RNASEH2C encodes a subunit of the RNase H2 enzyme complex, which is involved in DNA replication repair and importantly in innate immune sensing of nucleic acids. Mutations in RNASEH2C can cause autoinflammatory conditions due to accumulation of RNA/DNA hybrids that trigger immune responses. In context, hypomethylation of the RNASEH2C promoter in high-stress individuals (and fibromyalgia patients) might lead to overexpression of this enzyme. One could speculate that heightened RNase H2 activity might alter how DNA/RNA debris is cleared and modulate immune activation thresholds. Chronic psychological stress has been associated with immune system activation and even features of accelerated immune aging; thus, an epigenetic change in an immune-related gene like RNASEH2C is plausible as part of the stress response imprint. It aligns with the notion that fibromyalgia (and possibly chronic stress states) involve subtle immune dysregulation or heightened inflammatory signalling, even if not to the degree of an autoimmune disease.
The third locus, cg13348458, lies in an intergenic region on chromosome 6 with no well-annotated gene nearby. The function of this region is unclear – it could reside in a long-range regulatory element (enhancer/suppressor) or within a non-coding RNA. Its hypomethylation in both fibromyalgia and the high-stress group suggests it is a sensitive marker of systemic stress or pain-related physiology. Further work would be needed to identify if a gene is regulated by this locus (for instance, if it is an enhancer for a distant gene). It is notable that epigenome-wide studies of stress have found many intergenic or non-coding regions showing methylation changes, not just coding genes. Such regions might affect gene expression through chromatin interactions or by encoding regulatory RNAs.
Overall, our findings lend support to the idea that psychological stress can induce epigenetic changes in peripheral tissues (saliva DNA) that mirror those found in chronic pain conditions. This bolsters the theory of a biological continuum wherein chronic stress may contribute to the development or exacerbation of conditions like fibromyalgia. It also raises interesting possibilities for biomarkers: If a small panel of CpGs is sensitive to both chronic stress and fibromyalgia, it might serve as an indicator of an individual’s cumulative stress exposure or vulnerability to stress-related illnesses. The three-CpG “Epi Vitality” panel was originally proposed as a potential aid in fibromyalgia identification; our results suggest it could also be indicative of one’s stress level, or at least that stress plays a role in driving this epigenetic signature.
5. Conclusions
Individuals reporting very high levels of daily stress demonstrated significantly lower DNA methylation at a targeted trio of CpG sites in their saliva DNA, compared to individuals with minimal stress. These three CpGs – located in the promoters of AFAP1 and RNASEH2C and an intergenic region – were originally identified as hypomethylated in fibromyalgia patients, and our study shows a similar hypomethylation trend in the context of psychological stress. This finding suggests that chronic stress may induce epigenetic alterations that mirror those found in chronic pain conditions, highlighting a potential molecular link between stress and pain pathophysiology. While the effect size is modest, the consistency of direction with fibromyalgia’s epigenetic signature is notable. This work provides a proof-of-concept that an “epigenetic stress index” (such as the 3-CpG Epi Vitality panel) could serve as an objective indicator of chronic stress burden. Future research should validate these results in larger and diverse populations, examine the reversibility of methylation changes with stress reduction, and explore whether such epigenetic markers can predict or mediate health outcomes. Ultimately, integrating epigenetic measures with psychological assessments may improve our understanding of how life stress translates into biological risk, paving the way for better prevention and intervention strategies for stress-related diseases.
References
- Garrett Dee; Rebecca Ryznar; Colton Dee; Epigenetic Changes Associated with Different Types of Stressors and Suicide. Cells. 2023, 12, 1258.
- A Test for Fibromyalgia? Saliva DNA Methylation. encyclopedia.pub. Retrieved 2025-7-21
- Daniel Ciampi de Andrade; Mariana Maschietto; Ricardo Galhardoni; Gisele Gouveia; Thais Chile; Ana C. Victorino Krepischi; Camila S. Dale; André R. Brunoni; Daniella C. Parravano; Ana S. Cueva Moscoso; Irina Raicher; Helena H. S. Kaziyama; Manoel J. Teixeira; Helena P. Brentani; Epigenetics insights into chronic pain: DNA hypomethylation in fibromyalgia—a controlled pilot-study. Pain. 2017, 158, 1473-1480.
