The paper aims to advance automatedautomated prostate segmentation prostate segmentation on n T2‑weighted MRI by introducing a hybrid topological atlas‑based method. It leverages a collection of pre‑labeledpre‑labeled MRI atlas images MRI atlas images to capture anatomical variability and produce accurate prostate segmentations. The approach was evaluated on 30 T2‑weighted MRI scans. Automated contours were compared with manual expert segmentations using the Dice Similarity Coefficient (DSC). The method achieved high DSC values, indicating that it closely approximates expert-defined prostate boundaries. This research demonstrated good overlap between automated and manual segmentations, proving that the hybrid approach is effective and offered a quantitative performance benchmark: consistently solid DSC scores across the 30-case dataset.
Experimental Design
-
Data: The approach was evaluated on 30 T2‑weighted MRI scans.
-
Evaluation metric: Automated contours were compared with manual expert segmentations using the Dice Similarity Coefficient (DSC).
-
Outcome: The method achieved high DSC values, indicating that it closely approximates expert-defined prostate boundaries.
📊 Key Results
-
Demonstrated good overlap between automated and manual segmentations, proving that the hybrid approach is effective.
-
Offers a quantitative performance benchmark: consistently solid DSC scores across the 30-case dataset.
- MRI T2
- image segmentation
- prostate
- image
- segmentation
- atlas
- hybrid method
- medical informatics
1. 📌 Objective & Motivation
The method introduces a hybridhybrid topological atlas-based algorithm topological atlas-based algorithm for fully automatic prostate segmentation using T2-weighted MRI, designed to address limitations in time and accuracy of manual delineation. It uses a set of pre-labeled atlas images, aiming to deliver robust contours across patients with diverse prostate shapes.
2. 🌐 Atlas-Based & Topological Approach
Atlas Preparation
-
A small but diverse cohort of prostate MRIs are manually segmented to create labeled atlas images.
-
The atlas captures variations in size, shape, and orientation across prostates.
Matching & Registration
-
Leveraging the union of compatible contours from selected atlases, a composite “inferred prostate region” is built.
-
The method then maps these contours precisely to the target image, granting an automated prostate outline.
-
A new MRI (target) is aligned spatially to each atlas via image registration.
-
The topological similarity (contour shape, directional edges, intensity gradient) between atlases is computed.
-
This determines which atlas (or weighted combination) best matches the target’s topology.
Contour Estimation
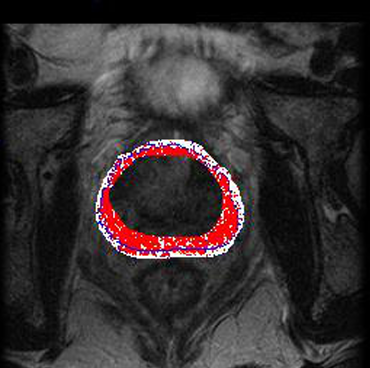
3. Performance & Evaluation
🧠 Summary Comparison Table
| Aspect | Topological Atlas Method |
|---|
-
T
-
Lestved on 30 T2-weighted prostate MRI scans, with segmentations compared to expertraging the manual delineations.
union of compatible contours from selected atlases, a composite “inferred prostate region” is built.
Evalua
-
The metehod via Dice Similarity Coefficient (DSC): results indicated high overlap, demonstrating strong segmentation accuracythen maps these contours precisely to the target image, granting an automated prostate outline.
-
The algorithm exhibited fast execution; typically, only a small region of interest is processed, reducing computational load.

3. ✅ Performance & Evaluation
-
Tested on 30 T2-weighted prostate MRI scans, with segmentations compared to expert manual delineations.
-
Evaluated via Dice Similarity Coefficient (DSC): results indicated high overlap, demonstrating strong segmentation accuracy.
-
The algorithm exhibited fast execution; typically, only a small region of interest is processed, reducing computational load.
4. 🎯 Strengths & Constraints
Strengths
| Modern Deep Learning Models | ||
|---|---|---|
| Data | 30 manually segmented MRIs | Hundreds to thousands of annotated MRIs |
| Approach | Atlas registration + contour union |
Computationally efficient, as only relevant subregions are processed.
Limitations
| CNNs (e.g., U-Net, V-Net), Transformers | ||
| Performance | Good DSC; efficient contours | Higher DSC (≈0.90 W.G., ≈0.79–0.87 zonal) |
| Speed | Quick due to ROI focus | Varies; commonly fast with GPU acceleration |
| Scalability | Atlas-limited; manual atlas prep | Scalable; needs large annotated datasets |
| Robustness | Sensitive to anatomy not seen in atlas | Generally robust with data augmentation |
-
Relies heavily on the atlas database: new prostate shapes not represented may be missed.
-
If the target’s unique anatomy (e.g., due to severe hypertrophy) lies outside the atlas union, segmentation may fail.
-
Topologically driven matching ensures segmentation respects prostate shape and contour direction.
-
It’s conceptually straightforward and easier to implement than complex deep learning methods.
-
Relies heavily on the atlas database: new prostate shapes not represented may be missed.
-
If the target’s unique anatomy (e.g., due to severe hypertrophy) lies outside the atlas union, segmentation may fail.
5. 📚 Broader Context & Follow-up Methods
Atlas vs. Deep Learning Today
-
Good fit for institutions with a small atlas archive aiming for quick deployment.
-
Modern approaches (e.g., U-Net, 3D CNNs, transformers) achieve DSC ≈ 0.9+ for whole gland; zonal segmentation lags slightly behind (DSC ≈ 0.79–0.87).
-
Data robustness is key: domain adaptation and boundary-aware losses help compensate for MRI variance and boundary uncertainty.
Technical Evolution
-
3D multi-planar CNNs (axial + sagittal scans) improved accuracy near prostate apex/base.
-
Transformer-based models now use cross-slice attention to better capture structural continuity.
-
3D multi-planar CNNs (axial + sagittal scans) improved accuracy near prostate apex/base.
-
Transformer-based models now use cross-slice attention to better capture structural continuity.
6.
7. Practical Implications
-
Suite for resource-limited environments: reproducible without massive datasets or specialized hardware.
-
Ideal as a baseline or teaching tool to understand topological segmentation before transitioning to neural methods.
7. 🧩 Practical Implications
-
Suite for resource-limited environments: reproducible without massive datasets or specialized hardware.
-
Good fit for institutions with a small atlas archive aiming for quick deployment.
-
Ideal as a baseline or teaching tool to understand topological segmentation before transitioning to neural methods.
8. 🔮 Future Directions
To enhance the topological method, one could:
-
Expand atlas diversity – more cases, including atypical anatomy.
-
Integrate ML-derived shape priors to handle contour variability absent in atlas.
-
Use hybrid pipelines: topological outputs guide or refine deep learning segmenters.
-
Blend active contour techniques to adapt atlas contours dynamically rather than fixed mapping.
-
Expand atlas diversity – more cases, including atypical anatomy.
-
Integrate ML-derived shape priors to handle contour variability absent in atlas.
-
Use hybrid pipelines: topological outputs guide or refine deep learning segmenters.
-
Blend active contour techniques to adapt atlas contours dynamically rather than fixed mapping.
9. 📝 In Summary
The topologicaltopological atlas-based method atlas-based method (2014) offers a fast,fast, interpretable, and implementation-friendly approach interpretable, and implementation-friendly approach for segmenting the prostate on T2 MRI. While deep learning now dominates—with higher accuracy, scalability, and anatomical generalizability—this method remains relevant for small-scale environments and algorithmic education. It underscores the persistent value of topology-driven segmentation, and provides foundational insights for modern hybrid systems that blend atlas priors with data-driven learning .
References
- Daniela Hristov; Done Stojanov; In silico report on five high-risk Protein C pathogenic variants: G403R, P405S, S421N, C238S, and I243T. Mutat. Res. Mol. Mech. Mutagen.. 2025, 831, 111907.
- Daniela Hristov; Done Stojanov; Exploring Regulatory Properties of Genes Associated with Nonsyndromic Male Infertility. Reprod. Med.. 2024, 5, 136-153.
