The genetic code symmetry is additional proofreading mechanism to prevent tRNA misreading in process of proteinogenesis. tRNA is a bridging molecule which leads to the identification of enzymes responsible for establishing a link between the nucleotides of mRNA and aminoacyl-tRNA synthetases (aaRSs).
- mRNA
- tRNA
- translation
- proteinogenesis
- genetic code symmetry
- the supersymmetry genetic code table
- Watson-Crick base pairing
- evolution
- mRNA, tRNA, aaRSs, translation, proteinogenesis, genetic code symmetry, the supersymmetry genetic code table, Watson-Crick base pairing, evolution
1. [1]Introduction
The group of enzymes aaRSs plays a crucial role in translation as an essential and universally distributed family of enzymes, pairing tRNA with their cognate amino acids for decoding mRNA according to the genetic code. Cognate amino acid corresponds to the anticodon triplet of the tRNA according to the genetic code. The aaRSs is the only enzyme capable of implementing the genetic code. Accordingly, the aaRSs prevents translation errors in the case of incorporating the wrong non-cognate amino acid using hydrolysis through deacetylation activity. As a result, the basal level of translational errors is about 10−4 amino acids.
Among the DNA molecule, genetic code, mRNA, tRNA and aaRSs, only the supersymmetry genetic code (SSyGC) table (1-3) as physicochemical symmetrically grouped codons for all individual amino acids including the sextets Serine, Arginine, and Leucine. Only one aaRSs is responsible for each amino acid. In this way, only aaRSs and the genetic code recognise all codons corresponding to a single amino acid [1].
2.
The Ssymmetries of Ccodons
However, due to the physicochemical similarities of certain amino acids, some aaRSs must rely on additional proofreading mechanisms to prevent tRNA misamnoacylation. These additional mechanisms are symmetries of the genetic code (Figure 1).
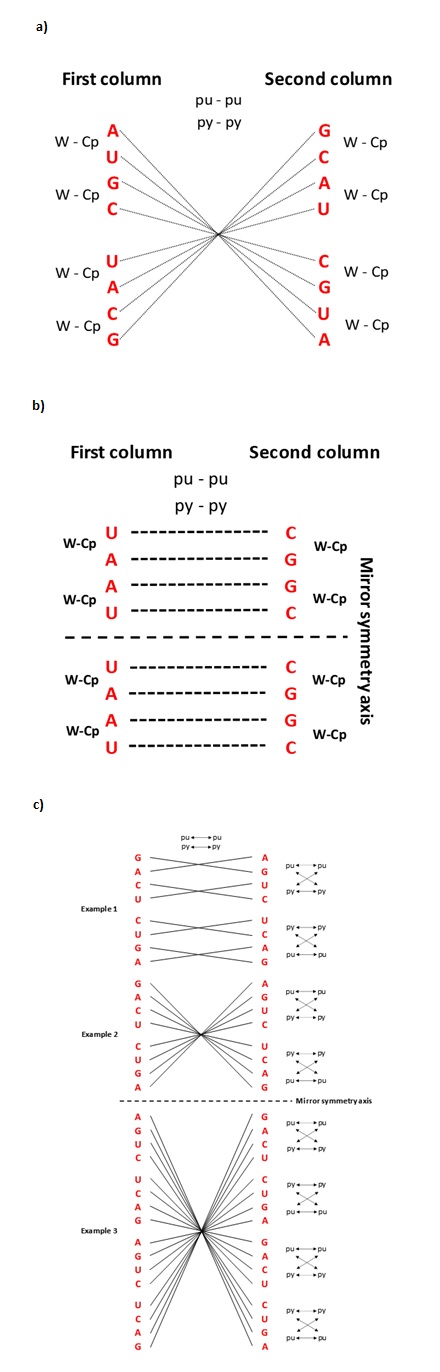
Figure 1
[2]Figure 1. Specific symmetries of each base from codons of the genetic code. (a) The centre point of mirror symmetry for the first base of codons in the SSyGC table. Equal first bases in the opposite pair of boxes on each symmetry line passing through the centre point. The first bases pointed the composing principle of codon boxes in the whole SSyGC table (in each box all four codons have identical first and second bases): first bases from a pair of boxes in a vertical direction have Watson-Crick pairing (W-Cp); the pair of boxes in a horizontal direction have purine-purine (pu-pu), pyrimidine-pyrimidine (py-py) pairing. (b) The second base of codons in the SSyGC table. Each pair of bases in the vertical direction are in the form of Watson-Crick base pairing (A↔U, C↔G), and in the horizontal direction are in purine—purine (G↔A) and pyrimidine—pyrimidine (C↔U) pairing. Both halves of code are identical according to the horizontal symmetry axis. It is very important to observe that the second bases in the whole left column are weak A and U, and in the right column are strong C and G. Due to this separation, the important rule of codon second base in differentiation of the amino acids with two or four codons in the flow from mRNA to tRNA is emphasised, which is important for the translation process especially wobbling pairing in proteinogenesis [1](1). (c) The centre points of cascade mirror symmetry for the third base of codons in the SSyGC table. All connecting lines between the same bases in the left and right columns passing through the centre point of the 1st, 2nd and 3rd example of mirror symmetry. Each cascade mirror symmetry comprises all third bases of the whole genetic code. Also, each pair of bases in the vertical and horizontal directions are in the form of purine—purine (G↔A) and pyrimidine—pyrimidine (C↔U) pairing. The third bases of the vertical pair of boxes have Watson-Crick pairing (G-A-C-U → C-U-G-A and A-G-U-C → U-C-A-G) and the newly discovered purine-pyrimidine symmetries of the SSyGC table. The Standard Genetic Code table ignored the third base of codons because all 16 boxes have an identical U-C-A-G alignment of the third base and cannot show Watson-Crick (codon—anticodon) symmetries between bases and codons [1][2](1).
3. The Ssymmetry Llanguage of Ccodons for Ttranscription
Due to symmetries, each codon has a strictly defined and unchangeable position within the genetic code. As a result, the first two bases in the codon reveal whether the codon belongs to a non-split box (all four codons belong to one amino acid), or to a split box which contains two amino acids, each with two codons.
According to the characteristics of the first two bases in codons in the SSyGC table, it can also distinguish four groups of codon boxes:
- non-split boxes with strong bases (GG, CC, GC, CG),
- split boxes with weak bases (AA, UU, AU, UA),
- mixed boxes with one strong and one weak base, which are not split because the second base is always pyrimidine (AC, UC, GU, CU),
- mixed boxes which are split because the second base is always purine (GA, CA, AG, UG) (Figure 2).

Figure 2
Figure 2. The first two bases determine the position of each codon from split or non-split boxes of amino acid with two or four codons in the genetic code. The characteristics of the first two bases from codons into each box: (a) strong (strg) CC, GG, CG, GC → non-split boxes because of both strong bases; (b) weak (wk) AA, UU, AU, UA → split boxes because of both weak bases; (c) a mix of weak and strong bases AC, UC, GU, CU → non-split boxes because the second base is pyrimidine; (d) a mix of weak and strong bases AG, UG, CA, GA → split boxes because the second base is purine. The sextets Serine, Arginine and Leucine include one non-split box (4/6) and a half of the neighbouring box with two codons (2/4) in the SSGC table. The purine-pyrimidine characteristics of the first two bases are very important for the differentiation of codons and translation from mRNA to anticodon tRNA. Namely, the first and second bases in mRNA always have canonical pairing in the process of proteinogenesis. Only the third base can have a wobble pairing. One amino acid with two codons from the same box of genetic code has a third base of purines A and G, and the oth[3]er amino acid pyrimidines U and C. The third base purines A or G have start/stop signals and Tryptophan with only one trinucleotide [1](1).
In conclusion, the symmetries of genetic code are not purely mathematical constructions but also represent physicochemical categories for differentiating codons of the SSyGC table on the principle of purine- pyrimidine Watson-Crick pairing. On the same principle of symmetries, the tRNA and aaRSs in process of translation can distinguish codons for its amino acid. It is very important to stress that the physicochemical purine-pyrimidine SSGC table is unique for all RNA and DNA species and unchangeable during evolution from the last universal common ancestor (LUCA) [1](Ref. 1) to Homo sapiens sapiens and such a way represent unique code of life.
References
- The Supersymmetry Genetic Code table. Scholarly Community Encyclopedia 2023.
- Rosandić, M.; Paar, V. Standard Genetic code vs. Supersymmetry Genetic Code – Alphabetical table vs. physicochemical table. . BioSystems. 2022, 14, 110748.
- Rosandić, M.; Paar, V. Maximal Genetic Code Symmetry is a Physicochemical Purine-Pyrimidine Symmetry Language for Transcription and Translation in the Flow of Genetic Information from DNA to Protein. Int. J. Mol. Sci. . 2024, 25, 9543.
