Microbiome research has revolutionized our understanding of complex microbial communities, and two pivotal techniques at the forefront of this exploration are 16S rRNA sequencing and metagenome sequencing.
- biotech
1. Introduction
Microbiome research has revolutionized our understanding of complex microbial communities, and two pivotal techniques at the forefront of this exploration are 16S rRNA sequencing and metagenome sequencing. These methodologies, each with unique strengths and applications, enable scientists to unravel the intricacies of microbial ecosystems. In this article, we delve deeper into the intricacies of these techniques and provide an in-depth guide to help researchers navigate the choice between 16S rRNA sequencing and metagenome sequencing based on their specific research objectives.
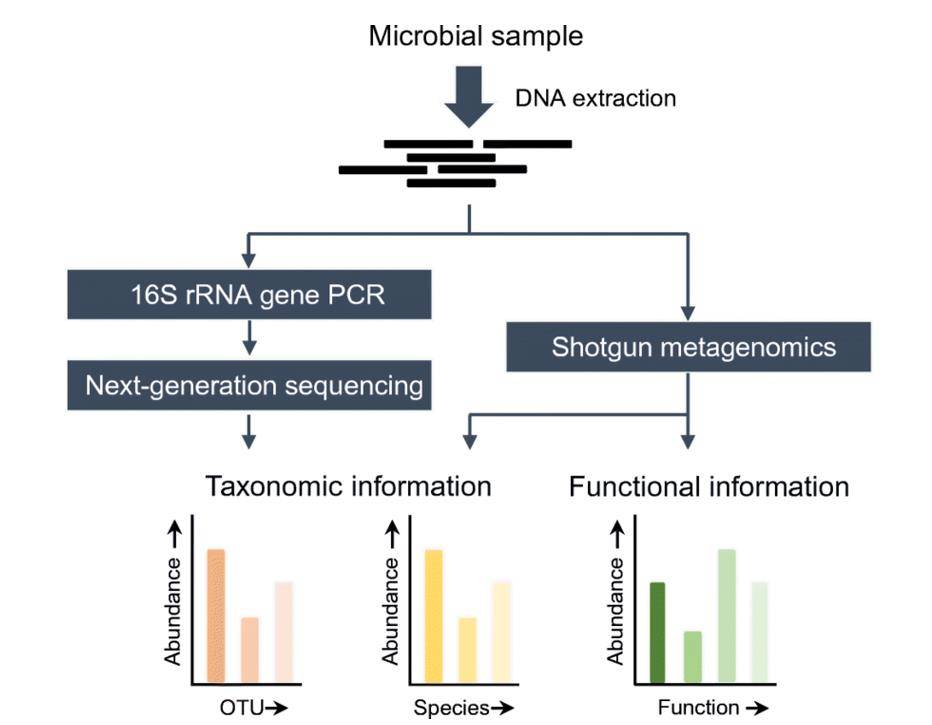 Overview of 16S rRNA gene NGS and shotgun metagenomics methods. (Boers et al., 2019)
Overview of 16S rRNA gene NGS and shotgun metagenomics methods. (Boers et al., 2019)
Overview of 16S rRNA gene NGS and shotgun metagenomics methods. (Boers et al., 2019)
2. 16S rRNA Sequencing
The 16S ribosomal DNA (rRNA) sequencing technique targets a segment within the small subunit of prokaryotic ribosomes, ranging from 1300 to 1600 base pairs in length. This region contains 10 conserved regions that reflect the evolutionary kinship between biological species, alongside 9 highly variable regions that highlight inter-species differences. The conserved regions act as a foundation for taxonomic identification, while the variable regions encapsulate unique distinctions among species. Typically, PCR amplification of a highly variable region (such as V4 or V3-V4) or the entire length of the 16S rRNA sequence is followed by sequencing.
3. Metagenome Sequencing
Metagenome sequencing transcends the boundaries of individual 16S rRNA sequences by encompassing all genetic material within a sample, including host DNA and microbial genomes. This all-encompassing approach grants insights into both taxonomic composition and functional potential. Through high-throughput sequencing of environmental DNA, metagenome sequencing facilitates analyses encompassing microbial community structure, gene content, and functional pathways.
4. Factors that Steer the Decision
When working with samples containing a well-defined set of known microorganisms, 16S rRNA sequencing proves to be adequate and fitting. In stark contrast, metagenome sequencing truly shines when faced with the exploration of intricate and diverse samples that house novel or poorly characterized species.
- Functional Insights
Metagenome sequencing empowers researchers with the capability to unveil intricate functional insights. This includes the ability to predict metabolic pathways and identify potential functional roles. Such revelations hold immense value, especially for studies that traverse the realms of ecology, medicine, and biotechnology.
5. 16S rRNA Sequencing vs. Metagenomic Sequencing
| Factors | 16S rRNA Sequencing | Metagenomic Sequencing |
| Genus (potentially species); influenced by targeted regions | ||
| Bacterial species (and possibly strains and single nucleotide variants with ample sequencing depth) | ||
| Taxonomic Coverage | Embraces bacteria and archaea | Encompasses all taxa, including viruses |
| Bioinformatics Requirements | Suited for those with beginner to intermediate expertise | Calls for intermediate to advanced proficiency |
| Databases | Draws from established, meticulously curated databases | Relies on relatively nascent and evolving databases |
| Sensitivity to Host DNA Contamination | Minimal impact (PCR success hinges on microbiome presence and absence of inhibitors) | Highly sensitive; subject to variation by sample type (adjustable by adapting sequencing depth) |
- Research Objectives
The articulation of precise research objectives holds utmost significance. When the intention revolves around deciphering aspects such as species composition, diversity, and relative abundance, the application of 16S rRNA sequencing becomes a judicious choice. In scenarios where investigations delve into granular aspects like specific functions, gene content, and intricate pathways, metagenome sequencing emerges as the favored and more appropriate approach.
- Taxonomic Resolution
16S rRNA sequencing excels in providing higher taxonomic precision, frequently extending down to the levels of genus or species. In contrast, metagenome sequencing delivers superior precision, enabling the discernment of subtle differences at the strain level, and remarkably, the potential reconstruction of complete genomes.
- Cost and Throughput
It is imperative to take into account financial constraints. 16S rRNA sequencing offers an economically viable and high-throughput solution, making it particularly advantageous when dealing with extensive community profiling endeavors. Conversely, metagenome sequencing entails higher costs due to the considerable volume of generated data and the associated computational requirements.
| Sample Preparation | ||
| Similar complexity to shotgun sequencing | Equally intricate as 16S rRNA sequencing | |
| Functional Profiling (Microbial Genes) | Potential for predicted functional profiling | Offers an extensive view of functional potential, though not definitively conclusive |
| Taxonomic Resolution | ||
| Bias | ||
| Moderate to high (taxonomic composition influenced by chosen primers and variable region) | ||
| Less pronounced (although impartial, potential biases can emerge during experimental and analytical phases) |
- Sample Complexity
Combining both methodologies can yield comprehensive insights. Utilize 16S rRNA sequencing for taxonomic profiling and initial community assessment, and subsequently employ metagenome sequencing to unravel functional potential and finer taxonomic nuances.
6. Conclusion
- Cost Constraints: Opt for 16S rRNA sequencing when budget is limited.
- Research Focus: For species composition and diversity, 16S rRNA sequencing suffices. Metagenome sequencing is essential for functional and gene-level inquiries.
- Genome Acquisition: Metagenome sequencing offers genome-level insights, even for uncultured bacteria.
- Hybrid Approach: Combine 16S rRNA sequencing with metagenome sequencing for cost-effective species-level studies alongside deeper functional analysis.
- Host Contamination: Choose 16S rRNA sequencing for samples with potential eukaryotic host DNA contamination.
7. Case Studies
Case Study 1 - Exploring the Bacterial Microbiome of Amazonian Soil
In this illustrative study, the target revolves around scrutinizing the intricate bacterial microbiome present in Amazonian soil. Given the circumstances elaborated earlier, where certain taxa might be rare or inadequately studied, opting for bacterial 16S rRNA databases proves advantageous. This choice stems from the lack of readily available full reference genomes for these specific species. Thus, leveraging 16S rRNA sequencing would offer enhanced taxonomic resolution compared to metagenomic sequencing, which aligns aptly with the study's objectives.
Case Study 2 - Tracking Microbiome Changes and Antimicrobial Gene Presence Post Fecal Transplant
The salient advantage of metagenomic sequencing lies in its capacity to furnish multifaceted functional microbiome insights, encompassing microbial gene data. For the scenario delineated here, a study aimed at deciphering shifts in microbiome composition and the carriage of antimicrobial genes subsequent to a fecal transplant, metagenomic sequencing emerges as the preferable choice. By adopting metagenomic sequencing, the investigation can effectively capture alterations in gut microbiome makeup and simultaneously delve into comprehensive profiling of antimicrobial resistance genes along with their associated hosts.
Case Study 3 - Daily Dynamics of Gut Microbiome Post 2-Week Dietary Fiber Intervention
Within the context of studying the daily oscillations in gut microbiome due to a 2-week dietary fiber intervention, certain instances arise where modifications manifest predominantly at the functional level. In such a study scenario, a judicious selection would be shallow shotgun sequencing. This approach uniquely empowers the assessment of both compositional variations (such as species or strain shifts) and functional disparities within the gut microbiome consequent to the dietary intervention. Notably, this method extends these analytical benefits at a comparable cost to that of 16S rRNA sequencing.
