Targeting the microbiome, microbiota-derived metabolites, and related pathways represents a significant challenge in oncology. Microbiome analyses have confirmed the negative impact of cancer treatment on gut homeostasis, resulting in acute dysbiosis and severe complications, including massive inflammatory immune response, mucosal barrier disruption, and bacterial translocation across the gut epithelium. Moreover, recent studies revealed the relationship between an imbalance in the gut microbiome and treatment-related toxicity. Recently, microbiota modulation via probiotic supplementation and fecal microbiota transplantation represents a new trend in cancer patient care, aiming to increase bacterial diversity, alleviate acute and long-term treatment-induced toxicity, and improve the response to various treatment modalities. A more detailed understanding of the complex relationship between the microbiome and host can significantly contribute to integrating a microbiome-based approach into clinical practice.
- the gut microbiome
- dysbiosis
- cancer treatment efficacy
- late effects
- cognitive impairment
- cardiotoxicity
- probiotics
- fecal microbiota transplantation
1. Introduction
2. Dominant Bacteria-Driven Mechanisms Associated with Cancer Development
The unmodifiable intrinsic and the modifiable or partially modifiable extrinsic factors affect cancer risk [8][9]. Studies showed that beyond microorganisms, numerous risk factors and their complex interplays, such as genetics and inherited mutations, geographical location, gender ratio, age, environmental exposure, and endogenous hormones, contribute to carcinogenesis within human populations [10][11]. A healthy lifestyle, diet, and nutrition play a key role in cancer prevention, and a higher-quality diet might reduce cancer risk [12]. On the other hand, unhealthy lifestyle and obesity are associated with cancer development [13]. Different dietary patterns significantly influence the composition of the gut microbiome [14]. Preparing a diet via the frying process might produce harmful carcinogenic acrylamide as a part of the Maillard reaction, negatively affecting gut microbiome homeostasis [15]. A comprehensive meta-analysis showed a lower incidence of cancer in vegetarians and vegans [16]. Similarly, Papadimitriou et al. performed an umbrella review of meta-analyses of observational studies to reveal associations between diet or nutrient intake and the risk of 11 primary cancers. Authors observed that dietary products, milk, and calcium were inversely associated with colorectal cancer (CRC), while drinking alcohol correlated positively with breast, colorectal, esophageal, liver, head, and neck malignancies [17]. Geographical provenance affects the composition of bacterial communities residing in the gastrointestinal tract [18]. Recently, an increasing number of studies confirmed that certain pathogenic microbes contribute to cancer development and progression via impact on DNA in host somatic cells, interrupted cell cycle, increased cell proliferation, and damaged processes responsible for apoptosis [19]. Almost 20% of all cancers might be related to microbial infection [20]. Specific bacteria produce toxins, leading to chronic inflammation with altered cellular processes [21]. However, not all infections with pathogenic microbes lead to cell malignant behavior and cancer development. Genetic heterogeneity of microorganisms and host genetics significantly affect cancer prevalence [22]. Unfavorable microbiota-derived metabolites could exhibit procarcinogenic properties and cause DNA double-strand breaks (DSBs) [23]. Conversely, beneficial bacterial metabolites might exert anti-cancer effects. Microbiota-derived short-chain fatty acids (SCFA) affect not only gut signaling pathways but also organs and tissues via blood circulation [24]. The main SCFA produced in the colon are acetate, propionate, and butyrate [25].2.1. Helicobacter pylori
Helicobacter pylori, first discovered by Barry Marshall and Robin Warren in 1984 [26], is a gram-negative micro-aerophilic bacterium that might be found in the upper intestinal tract within 50% of the population worldwide [27]. Possible routes of bacterial transmission are via saliva or feces [28]. The colonization of the gastric mucus layer by Helicobacter pylori is mediated via adhesins that bind Lewis determinants and mucin 5 (MUC5AC) [29][30]. Bacterial strains can be either cytotoxin-associated gene A (cagA) positive or cagA negative due to inserted cag pathogenicity island (cagPAI) containing approximately 32 genes [31]. Infection with Helicobacter pylori promotes DNA DSBs and induces host genomic instability. The accumulation of DSBs was higher in the case of cagPAI-positive bacterial strains, and the presence of a cagPAI can double the risk of gastric cancer incidence [32]. Within cagPAI, microsyringe (needle-like pilus) coded genes formed the Type IV secretion system (T4SS), which plays a role in CagA oncoprotein translocation into gastric epithelial cells [33]. Oncoprotein is responsible for disrupted epithelial tight junctions and lost apical-basolateral polarity in cells via CagA interaction with partitioning-defective 1 (PAR1)/microtubule affinity-regulating kinase (MARK). CagA prevents PAR1 phosphorylation mediated by atypical protein kinase C (aPKC), resulting in PAR1 dissociation from the membrane [34].2.2. Fusobacterium nucleatum
Fusobacterium nucleatum is an anaerobic bacterium residing in the human gut and oral microbiome, where it co-exists with other microorganisms [35]. In 2012, two individual studies found enrichment of Fusobacterium nucleatum in CRC compared to adjacent tissue samples via RNA and whole-genome sequencing [36][37]. This bacterium is linked not only to CRC but also to other human diseases such as periodontal diseases, dental pulp infections, halitosis, oral cancer, infections of the respiratory tract, appendicitis, cardiovascular disease, pregnancy disorders, breast cancer, and rheumatoid arthritis [38][39][40]. FadA and Fap2 represent the key virulence factors of Fusobacterium nucleatum. The role of FadA is mediating the attachment and binding to host cells [41]. FadA binds to the specific binding site of E-cadherin and causes bacterial invasion into host epithelial cells [42]. Specific mechanisms of how Fusobacterium nucleatum supports inflammation and CRC tumorigenesis result from FadA-induced activation of the Wnt/β-catenin signaling pathway [42][43].2.3. Escherichia coli
Escherichia coli represents another potential pathogen implicated in CRC. The studies showed a higher prevalence of enteropathogenic Escherichia coli (EPEC) in CRC patients compared to healthy controls. Moreover, Escherichia coli in patients serotypically and genotypically differed from those in the general population [44]. Pathogenic strains produce toxins, including colibactin, cytolethal distending toxin (CDT), cycle inhibiting factor, and cytotoxic necrotizing factor [45]. CDT is a genotoxin composed of CdtA, CdtB, and CdtC subunits [46], while the CdtB catalytic subunit might support carcinogenesis and host cell transformation in murine experiments [47]. The results from clinical studies showed that colibactin coded by Pks island was observed mainly in CRC patients [45].2.4. Salmonella
Microbial products of typhoid toxin and toxins like nitroso-chemical compounds produced by Salmonella typhi might be responsible for the potential development of tumors on the side of infection. The infection with Salmonella Paratyphi A caused DNA damage in gallbladder organoids. The experimental results supported associations between Salmonella, epithelial cell invasion, initiating malignant transformation, and gallbladder carcinogenesis [48]. Moreover, severe bacterial infection with Salmonella might contribute to CRC development [49][50]. Salmonella secreted AvrA, a multifunctional protein that activates Wnt and STAT3 signaling pathways, resulting in enhanced proliferation of CRC cells [51]. In vivo experiments showed that AvrA regulates several other pathways, including mTOR, NFκB, oxidative phosphorylation, platelet-derived growth factors, vascular endothelial growth factor, and mitogen-activated protein kinase signaling pathway [52]. AvrA inhibits macrophage death, leading to innate immune signaling blockade. Therefore, AvrA might establish a stable niche for intracellular Salmonella where the pathogen avoids adaptive immune responses [53].2.5. Bacteroides fragilis
The enteric pathogen known as enterotoxigenic Bacteroides fragilis (ETBF) secretes toxin (BFT) coded by the bft gene [54]. ETBF strains are implicated mainly in acute diarrheal diseases, but the studies highlight the microbe’s participation in CRC [55]. In vitro experiments documented that BFT increased the production of ROS, induced DNA damage, and promoted tumorigenesis [55]. This toxin is responsible for the damaged epithelial barrier and activated STAT3/Th17 immune responses [56][57].2.6. Staphylococcus aureus
This gram-positive bacterium produces several toxins and virulence factors, including Staphylococcal enterotoxin A (SEA) and Staphylococcal enterotoxin B (SEB) [58]. In most cases, Staphylococcus aureus is responsible for developed bacteremias in patients with hematologic malignancies [59][60]. Acute myeloid leukemia (AML) cell line revealed increased proliferation after treatment with SEA and SEB virulence factors in vitro [61]. In contrast, an experimental study on glioblastoma cells showed that SEB reduced smad2/3 expression and decreased cancer cell proliferation [62].2.7. Campylobacter jejuni
The presence of Campylobacter is associated with inflammation and implicated in the activation of mTOR signaling and neutrophil infiltration [63]. Campylobacter colonizes the intestinal tract due to adherence of Campylobacter jejuni CadF and FlpA adhesins to fibronectin. Both adhesins are responsible for physical contact with host cells, contributing to bacterial adherence, invasion, and cell signaling [64]. CDT produced by Campylobacter induces DNA damage via DSBs.2.8. Desulfovibrio
Desulfovibrio, belonging to sulfate-reducing bacteria (SRB), might participate in CRC development via hydrogen sulfide (H2S) production. Higher concentration of H2S damages DNA, leading to genomic and chromosomal instability [65][66]. Kapral et al. found that LPS from Desulfovibro desulfuricans altered the activity of p65 and IκBα genes in Caco-2 colon cancer cells [67]. Moreover, Desulfovibrio abundance was significantly higher in patients with advanced gastric cancer (stage IV).2.9. Porphyromonas
Recently, the results from several studies proposed that Porphyromonas gingivalis, as an oral pathogen, is implicated in pancreatic and oral tumorigenesis [68][69]. Porphyromonas LPS increased gingival stem/progenitor cell proliferation [70]. Gingipains (proteases) secreted by Porphyromonas gingivalis are involved in oral cell Notch-1 activation and PLA2-IIA production [71].3. Microbiome and Treatment Efficacy
In cancer patients, several factors influence the gut microbiome composition. In addition to the malignant disease and the impact of genetic-, diet- and lifestyle-related factors, the administration of antibiotics, immunosuppressants, supportive agents, and especially anti-cancer treatment play a role. Chemotherapy, similar to radiotherapy, heavily disrupts the balance in the microbial environment and leads to gut dysbiosis (Figure 1).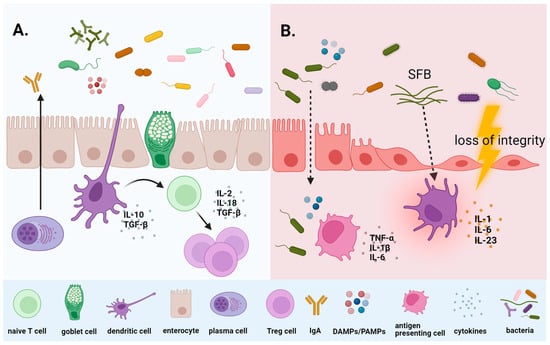
3.1. Microbiome and Chemotherapy
3.2. Microbiome and Immunotherapy
Immunotherapy represents a major advance in the clinical management of several cancers. Over the last decade, it has revolutionized the treatment of solid and hematologic malignancies, even those associated with a poor prognosis. The most widely used are immune checkpoint inhibitors (ICI), developed to enhance the activity of the body’s own immune cells against cancer cells [80]. ICI includes monoclonal antibodies designed to block the immune regulators, CTLA-4 (ipilimumab, tremelimumab), programmed death-1 (PD-1) (nivolumab, pembrolizumab), and PD-L1 (atezolizumab, avelumab, durvalumab) expressed in the cancer cells, with the consequent cytotoxic immune response. While these immunotherapies have improved patient outcomes in many clinical settings, they can induce toxicity, specifically immune-related adverse events. Commonly experienced adverse effects include cutaneous, musculoskeletal, intestinal, endocrine, and pulmonary, while cardiovascular, hematologic, renal, and neurological occur much less frequently [81]. However, the cardiovascular toxicity of ICI is of particular concern, given their impact on the morbidity and mortality of cancer patients [82]. Myocarditis is a severe complication of ICI with a high fatality rate that most frequently develops during the first 12 weeks of treatment, although late cases may occur [83][84]. Currently, an increasing number of studies are addressing the impact of the microbiome on the efficacy of immunotherapy, and accumulating evidence confirms that modulating the gut microbiome in favor of a favorable composition proves to be a promising trend in improving the response to immunotherapy using ICI [85]. The introduction of immunotherapy represents a key step in cancer treatment, with blocking CTLA-4, PD-1, and PD-L1 checkpoint pathways helping to restore the anti-tumor immune response [86]. The fundamental mechanisms underlying the relationship between the microbiome and the response to immunotherapy include increased infiltration of tumor immune cells, maturation of dendritic cells, and the production of IL-12, which promotes increased differentiation of Th cells and immune activation in the tumor microenvironment. It also involves the expansion of cytotoxic CD8 cells associated with the upregulation of perforin and serine protease granzyme, leading to apoptotic destruction of tumor cells [87][88].4. Microbiome and Therapy-Induced Late Effects
Cancer treatment, especially chemotherapy, causes a range of late complications in survivors, including neurological, ophthalmological, pneumatological, cardiological, and nephrological complications or issues linked to infertility and necrosis of the femoral head [89]. Cancer survivors experience disruption of the immune system correlated with therapy or the malignant disease. Considering that gut microbiome composition is crucial for shaping the immune system, the associations between the gut microbiome and treatment-induced late effects are gaining attention.4.1. Microbiome and Treatment-Induced Cognitive Impairment
The brain is highly sensitive to microbial disharmony, and the altered composition of the intestinal microbiome significantly affects the physiology and functions of the nervous system. Changes in the microbiota-host relationship affect the enteric nervous system and activate neuroimmune signaling pathways, influencing brain development and functioning [90]. Microbial signals, including structural bacterial components or microbiota-derived metabolites, can influence distant organs directly or through neural and hormonal signaling. Systemic inflammation induced by intestinal dysbiosis can increase the stress-activated “hypothalamus-pituitary-adrenal” (HPA) axis [91]. Mechanistic studies have revealed pathways through which communication occurs along the microbiome-gut-brain axis. The gut microbiota produces microbiota-derived metabolites such as SCFA, trimethylamine N-oxide (TMAO), endotoxins, and amino acids, circulating in the blood to the brain and affecting nervous functions. In addition to the role of SCFA in maintaining the integrity of the intestinal membrane and mucin production, SCFA´s involvement in signaling between the microbiome, gut, and brain via immune, endocrine, and humoral pathways is intensively studied [92]. Certain strains of gut bacteria can secrete neurotransmitters such as acetylcholine, gamma-aminobutyric acid (GABA), tryptophan, and serotonin. GABA is a neurotransmitter that helps to maintain the healthy functioning of the brain and nervous system [93]. Metagenomic and metabolomic analyses showed that not only higher levels of Fusobacteium nucleatum but also reduced SCFA and decreased GABA biosynthesis were implicated in late-onset CRC [94]。 Cancer treatment can lead to cognitive impairment associated with memory deficits, attention problems, information processing, and decision-making abilities. These negative impacts can persist long-term after the end of treatment, significantly affecting the lives of survivors. A comparison of cognitive functions in 581 breast cancer patients and 364 healthy individuals in the control group revealed that more than one-third of patients in the chemotherapy group experienced cognitive dysfunction that persisted for at least 6 months post-treatment [95]. The relationship between chemotherapy, radiotherapy, and decreased cognitive functions was also confirmed in a cohort of 155 testicular cancer patients [96].4.2. Microbiome and Cardiovascular Toxicity
The expanding range of cancer therapeutics has led to a broad spectrum of cardiovascular complications diagnosed in patients during and after cancer therapy. Moreover, high cardiotoxicity is the reason for treatment discontinuation. Cancer therapy-related cardiovascular toxicity includes cardiomyopathy, heart failure, myocarditis, coronary artery disease, peripheral vascular disease, hypertension, arrhythmias, pericardial, valvular heart diseases, and thromboembolism [97][98][99][100]. These complications are linked to chemotherapy (such as anthracycline cytostatics and platinum derivates), targeted agents (monoclonal antibodies and tyrosine kinase inhibitors), immunotherapy (including mainly ICI), and radiation therapy (to the left chest or mediastinum). Significant excesses in mortality risk associated with treatment-related complications, including cardiac causes, exist up to 2 years after the initial cancer diagnosis [101]. The relative risk of both arterial and venous thromboembolism is significantly higher in cancer patients compared with the general population [102]. A recent study comprising 12,414 ARIC (Atherosclerosis Risk In Communities) participants monitored for decades showed that cancer patients had a 52% higher risk of heart failure and a 22% higher risk of sudden stroke compared to patients without a cancer diagnosis [103]. TMAO arising from intestinal microbiota is a novel biomarker linked to atherosclerosis and risk of major adverse cardiovascular disease events and death in animals and humans [104][105][106][107]. TMAO levels have also been shown to correlate with pro-inflammatory state [108]. Due to its connection to dietary intake, TMAO could be influenced by intermittent fasting, and its change highlights the possibility that fasting may also beneficially alter the microbiome, at least during caloric restriction, if not for a more extended period of time after the completion of fasting. Benefits on metabolic health parameters, lower risk of coronary heart disease and depression, and cognitive performance improvement may be reached by a 24-hour water-only fasting intervention in apparently healthy individuals [109].5. Microbiota Modulation by Probiotics, Prebiotics, and Fecal Microbiota Transplantation in Cancer Patients
Mounting evidence highlights the emerging role of gut microbiota modulation in cancer patients via probiotic and prebiotic administration [110]. Fecal microbiota transplantation (FMT) from a healthy donor or treatment-responding patient quantitatively and qualitatively surpasses the supplementation with probiotics alone. The safety and efficacy of microbiota modulation in immunosuppressed cancer patients are the subject of intense research, and studies confirm the positive effect of modulation on patient outcomes (Figure 2).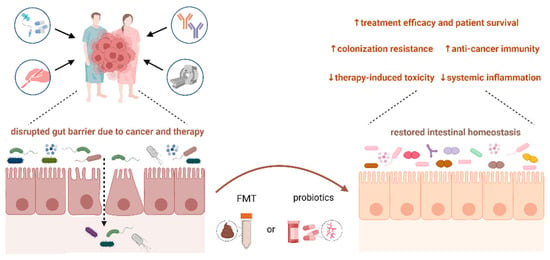
6. Critical Analysis of the Clinical Utility of a Microbiome-Based Approach
67. Conclusions
The role of the gut microbiome in cancer patients is not yet fully elucidated, but its importance is continually confirmed. Advances in complex molecular biology and genetic approaches, along with the development of sophisticated bioinformatic algorithms, have enabled extensive microbial analyses and brought us closer to understanding the true impact of the microbiome on human health. However, microbiome research is closely associated with several challenges, especially standardizing technological procedures for sample collection, including storage and sample processing. It also involves complex analysis of sequencing data and defining causal relationships between changes in microbiome composition and malignant diseases. Microorganisms can contribute to the initiation and progression of malignancies at both local and systemic levels by influencing the host immune response and producing metabolites and genotoxins by individual bacterial taxa. Clinical studies have confirmed extensive changes in the gut microbiome after undergoing anti-tumor therapy, with the most data available for patients treated with chemotherapy, radiotherapy, and immunotherapy. Simultaneously, the individual composition of the microbiome can activate the immune system and enhance the patient´s response to the administered treatment. Restoring Pthe balance in the gastrointestinal tract is possible in several ways, opening the opportunity for modulating the gut microbiota to reduce acute and late treatment toxicity and improve therapeutic response. Given the interaction between the gut and tumor microbiome, modulations may also influence the composition of the tumor microbiome. Therefore, personalized determination of the gut and tumor microbiome may represent a potential diagnostic and prognostic tool, and research in the coming years will reveal the most effective and safest ways to modify the microbiome to improve patient outcomes. Despite many challenges, it is highly likely that microbiome research in oncology will increasingly contribute to the diagnosis of cancer and the stratification of patients for the development of more effective, individualized, tumor-specific therapies in the next decade. The implementation of metatranscriptomic and metabolomic approaches, which complement metagenomic analyses, will also be of the highest interest. Importantly, machine learning algorithms might play a significant role, helping to uncover signaling networks for identifying new targets to predict treatment response.References
- Ramirez-Labrada, A.G.; Isla, D.; Artal, A.; Arias, M.; Rezusta, A.; Pardo, J.; Galvez, E.M. The Influence of Lung Microbiota on Lung Carcinogenesis, Immunity, and Immunotherapy. Trends Cancer 2020, 6, 86–97.
- Ciernikova, S.; Sevcikova, A.; Stevurkova, V.; Mego, M. Tumor microbiome—An integral part of the tumor microenvironment. Front. Oncol. 2022, 12, 1063100.
- Chen, X.; Song, M.; Zhang, B.; Zhang, Y. Reactive Oxygen Species Regulate T Cell Immune Response in the Tumor Microenvironment. Oxid. Med. Cell. Longev. 2016, 2016, 1580967.
- Kennel, K.B.; Greten, F.R. Immune cell—Produced ROS and their impact on tumor growth and metastasis. Redox. Biol. 2021, 42, 101891.
- Ciernikova, S.; Mego, M.; Hainova, K.; Adamcikova, Z.; Stevurkova, V.; Zajac, V. Modification of microflora imbalance: Future directions for prevention and treatment of colorectal cancer? Neoplasma 2015, 62, 345–352.
- Goubet, A.G. Could the tumor-associated microbiota be the new multi-faceted player in the tumor microenvironment? Front. Oncol. 2023, 13, 1185163.
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980.
- Wu, S.; Zhu, W.; Thompson, P.; Hannun, Y.A. Evaluating intrinsic and non-intrinsic cancer risk factors. Nat. Commun. 2018, 9, 3490.
- Wu, S.; Powers, S.; Zhu, W.; Hannun, Y.A. Substantial contribution of extrinsic risk factors to cancer development. Nature 2016, 529, 43–47.
- Mbemi, A.; Khanna, S.; Njiki, S.; Yedjou, C.G.; Tchounwou, P.B. Impact of Gene-Environment Interactions on Cancer Development. Int. J. Environ. Res. Public Health 2020, 17, 8089.
- Dorak, M.T.; Karpuzoglu, E. Gender differences in cancer susceptibility: An inadequately addressed issue. Front. Genet. 2012, 3, 268.
- Narimatsu, H.; Yaguchi, Y.T. The Role of Diet and Nutrition in Cancer: Prevention, Treatment, and Survival. Nutrients 2022, 14, 3329.
- Mittelman, S.D. The Role of Diet in Cancer Prevention and Chemotherapy Efficacy. Annu. Rev. Nutr. 2020, 40, 273–297.
- Ciernikova, S.; Sevcikova, A.; Stevurkova, V.; Mego, M. Diet-driven microbiome changes and physical activity in cancer patients. Front. Nutr. 2023, 10, 1285516.
- Shi, B.; Guo, X.; Liu, H.; Jiang, K.; Liu, L.; Yan, N.; Farag, M.A.; Liu, L. Dissecting Maillard reaction production in fried foods: Formation mechanisms, sensory characteristic attribution, control strategy, and gut homeostasis regulation. Food Chem. 2023, 438, 137994.
- Dinu, M.; Abbate, R.; Gensini, G.F.; Casini, A.; Sofi, F. Vegetarian, vegan diets and multiple health outcomes: A systematic review with meta-analysis of observational studies. Crit. Rev. Food Sci. Nutr. 2017, 57, 3640–3649.
- Papadimitriou, N.; Markozannes, G.; Kanellopoulou, A.; Critselis, E.; Alhardan, S.; Karafousia, V.; Kasimis, J.C.; Katsaraki, C.; Papadopoulou, A.; Zografou, M.; et al. An umbrella review of the evidence associating diet and cancer risk at 11 anatomical sites. Nat. Commun. 2021, 12, 4579.
- Fontana, A.; Panebianco, C.; Picchianti-Diamanti, A.; Lagana, B.; Cavalieri, D.; Potenza, A.; Pracella, R.; Binda, E.; Copetti, M.; Pazienza, V. Gut Microbiota Profiles Differ among Individuals Depending on Their Region of Origin: An Italian Pilot Study. Int. J. Environ. Res. Public Health 2019, 16, 4065.
- Whisner, C.M.; Athena Aktipis, C. The Role of the Microbiome in Cancer Initiation and Progression: How Microbes and Cancer Cells Utilize Excess Energy and Promote One Another’s Growth. Curr. Nutr. Rep. 2019, 8, 42–51.
- De Martel, C.; Ferlay, J.; Franceschi, S.; Vignat, J.; Bray, F.; Forman, D.; Plummer, M. Global burden of cancers attributable to infections in 2008: A review and synthetic analysis. Lancet Oncol. 2012, 13, 607–615.
- Vidal, M.C.; Anneberg, T.J.; Cure, A.E.; Althoff, D.M.; Segraves, K.A. The variable effects of global change on insect mutualisms. Curr. Opin. Insect Sci. 2021, 47, 46–52.
- Bhatt, A.P.; Redinbo, M.R.; Bultman, S.J. The role of the microbiome in cancer development and therapy. CA Cancer J. Clin. 2017, 67, 326–344.
- Yang, Q.; Wang, B.; Zheng, Q.; Li, H.; Meng, X.; Zhou, F.; Zhang, L. A Review of Gut Microbiota-Derived Metabolites in Tumor Progression and Cancer Therapy. Adv. Sci. 2023, 10, e2207366.
- Cong, J.; Zhou, P.; Zhang, R. Intestinal Microbiota-Derived Short Chain Fatty Acids in Host Health and Disease. Nutrients 2022, 14, 1977.
- Portincasa, P.; Bonfrate, L.; Vacca, M.; De Angelis, M.; Farella, I.; Lanza, E.; Khalil, M.; Wang, D.Q.; Sperandio, M.; Di Ciaula, A. Gut Microbiota and Short Chain Fatty Acids: Implications in Glucose Homeostasis. Int. J. Mol. Sci. 2022, 23, 1105.
- Marshall, B.J.; Warren, J.R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1984, 1, 1311–1315.
- Amieva, M.; Peek, R.M., Jr. Pathobiology of Helicobacter pylori-Induced Gastric Cancer. Gastroenterology 2016, 150, 64–78.
- Brown, L.M. Helicobacter pylori: Epidemiology and routes of transmission. Epidemiol. Rev. 2000, 22, 283–297.
- Chmiela, M.; Kupcinskas, J. Review: Pathogenesis of Helicobacter pylori infection. Helicobacter 2019, 24 (Suppl. S1), e12638.
- Gonciarz, W.; Walencka, M.; Moran, A.P.; Hinc, K.; Obuchowski, M.; Chmiela, M. Upregulation of MUC5AC production and deposition of LEWIS determinants by HELICOBACTER PYLORI facilitate gastric tissue colonization and the maintenance of infection. J. Biomed. Sci. 2019, 26, 23.
- Noto, J.M.; Peek, R.M., Jr. The Helicobacter pylori cag Pathogenicity Island. Methods Mol. Biol. 2012, 921, 41–50.
- Hanada, K.; Uchida, T.; Tsukamoto, Y.; Watada, M.; Yamaguchi, N.; Yamamoto, K.; Shiota, S.; Moriyama, M.; Graham, D.Y.; Yamaoka, Y. Helicobacter pylori infection introduces DNA double-strand breaks in host cells. Infect. Immun. 2014, 82, 4182–4189.
- Backert, S.; Tegtmeyer, N.; Fischer, W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015, 10, 955–965.
- Saadat, I.; Higashi, H.; Obuse, C.; Umeda, M.; Murata-Kamiya, N.; Saito, Y.; Lu, H.; Ohnishi, N.; Azuma, T.; Suzuki, A.; et al. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature 2007, 447, 330–333.
- Sun, C.H.; Li, B.B.; Wang, B.; Zhao, J.; Zhang, X.Y.; Li, T.T.; Li, W.B.; Tang, D.; Qiu, M.J.; Wang, X.C.; et al. The role of Fusobacterium nucleatum in colorectal cancer: From carcinogenesis to clinical management. Chronic Dis. Transl. Med. 2019, 5, 178–187.
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306.
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298.
- Chen, Y.; Huang, Z.; Tang, Z.; Huang, Y.; Huang, M.; Liu, H.; Ziebolz, D.; Schmalz, G.; Jia, B.; Zhao, J. More Than Just a Periodontal Pathogen-the Research Progress on Fusobacterium nucleatum. Front. Cell. Infect. Microbiol. 2022, 12, 815318.
- Liu, H.; Liu, Y.; Liu, W.; Zhang, W.; Xu, J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat. Commun. 2015, 6, 8494.
- Gaba, F.I.; Gonzalez, R.C.; Martinez, R.G. The Role of Oral Fusobacterium nucleatum in Female Breast Cancer: A Systematic Review and Meta-Analysis. Int. J. Dent. 2022, 2022, 1876275.
- Han, Y.W.; Ikegami, A.; Rajanna, C.; Kawsar, H.I.; Zhou, Y.; Li, M.; Sojar, H.T.; Genco, R.J.; Kuramitsu, H.K.; Deng, C.X. Identification and characterization of a novel adhesin unique to oral fusobacteria. J. Bacteriol. 2005, 187, 5330–5340.
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206.
- Guo, P.; Tian, Z.; Kong, X.; Yang, L.; Shan, X.; Dong, B.; Ding, X.; Jing, X.; Jiang, C.; Jiang, N.; et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J. Exp. Clin. Cancer Res. 2020, 39, 202.
- Magdy, A.; Elhadidy, M.; Abd Ellatif, M.E.; El Nakeeb, A.; Abdallah, E.; Thabet, W.; Youssef, M.; Khafagy, W.; Morshed, M.; Farid, M. Enteropathogenic Escherichia coli (EPEC): Does it have a role in colorectal tumourigenesis? A Prospective Cohort Study. Int. J. Surg. 2015, 18, 169–173.
- Villeger, R.; Lopes, A.; Veziant, J.; Gagniere, J.; Barnich, N.; Billard, E.; Boucher, D.; Bonnet, M. Microbial markers in colorectal cancer detection and/or prognosis. World J. Gastroenterol. 2018, 24, 2327–2347.
- Scott, D.A.; Kaper, J.B. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 1994, 62, 244–251.
- Tremblay, W.; Mompart, F.; Lopez, E.; Quaranta, M.; Bergoglio, V.; Hashim, S.; Bonnet, D.; Alric, L.; Mas, E.; Trouche, D.; et al. Cytolethal Distending Toxin Promotes Replicative Stress Leading to Genetic Instability Transmitted to Daughter Cells. Front. Cell Dev. Biol. 2021, 9, 656795.
- Sepe, L.P.; Hartl, K.; Iftekhar, A.; Berger, H.; Kumar, N.; Goosmann, C.; Chopra, S.; Schmidt, S.C.; Gurumurthy, R.K.; Meyer, T.F.; et al. Genotoxic Effect of Salmonella Paratyphi A Infection on Human Primary Gallbladder Cells. mBio 2020, 11, e01911-20.
- Duijster, J.W.; Hansen, J.V.; Franz, E.; Neefjes, J.J.C.; Frisch, M.; Mughini-Gras, L.; Ethelberg, S. Association between Salmonella infection and colon cancer: A nationwide registry-based cohort study. Epidemiol. Infect. 2021, 149, e56.
- Mughini-Gras, L.; Schaapveld, M.; Kramers, J.; Mooij, S.; Neefjes-Borst, E.A.; Pelt, W.V.; Neefjes, J. Increased colon cancer risk after severe Salmonella infection. PLoS ONE 2018, 13, e0189721.
- Lu, R.; Wu, S.; Zhang, Y.G.; Xia, Y.; Zhou, Z.; Kato, I.; Dong, H.; Bissonnette, M.; Sun, J. Salmonella Protein AvrA Activates the STAT3 Signaling Pathway in Colon Cancer. Neoplasia 2016, 18, 307–316.
- Liu, X.; Lu, R.; Xia, Y.; Wu, S.; Sun, J. Eukaryotic signaling pathways targeted by Salmonella effector protein AvrA in intestinal infection in vivo. BMC Microbiol. 2010, 10, 326.
- Wu, H.; Jones, R.M.; Neish, A.S. The Salmonella effector AvrA mediates bacterial intracellular survival during infection in vivo. Cell. Microbiol. 2012, 14, 28–39.
- Viljoen, K.S.; Dakshinamurthy, A.; Goldberg, P.; Blackburn, J.M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS ONE 2015, 10, e0119462.
- Pierce, J.V.; Bernstein, H.D. Genomic Diversity of Enterotoxigenic Strains of Bacteroides fragilis. PLoS ONE 2016, 11, e0158171.
- Yu, L.C.; Wei, S.C.; Ni, Y.H. Impact of microbiota in colorectal carcinogenesis: Lessons from experimental models. Intest. Res. 2018, 16, 346–357.
- Bundgaard-Nielsen, C.; Baandrup, U.T.; Nielsen, L.P.; Sorensen, S. The presence of bacteria varies between colorectal adenocarcinomas, precursor lesions and non-malignant tissue. BMC Cancer 2019, 19, 399.
- Pinchuk, I.V.; Beswick, E.J.; Reyes, V.E. Staphylococcal enterotoxins. Toxins 2010, 2, 2177–2197.
- Espersen, F.; Frimodt-Moller, N.; Rosdahl, V.T.; Jessen, O.; Faber, V.; Rosendal, K. Staphylococcus aureus bacteremia in patients with hematological malignancies and/or agranulocytosis. Acta Medica Scand 1987, 222, 465–470.
- Velasco, E.; Thuler, L.C.; Martins, C.A.; Nucci, M.; Dias, L.M.; Goncalves, V.M. Epidemiology of bloodstream infections at a cancer center. Sao Paulo Med. J. 2000, 118, 131–138.
- Turk, S.; Yanpar, H.; Baesmat, A.S.; Canli, S.D.; Cinar, O.E.; Malkan, U.Y.; Turk, C.; Haznedaroglu, I.C.; Ucar, G. Enterotoxins A and B produced by Staphylococcus aureus increase cell proliferation, invasion and cytarabine resistance in acute myeloid leukemia cell lines. Heliyon 2023, 9, e19743.
- Akbari, A.; Farahnejad, Z.; Akhtari, J.; Abastabar, M.; Mobini, G.R.; Mehbod, A.S. Staphylococcus aureus Enterotoxin B Down-Regulates the Expression of Transforming Growth Factor-Beta (TGF-beta) Signaling Transducers in Human Glioblastoma. Jundishapur J. Microbiol. 2016, 9, e27297.
- Sun, X.; Threadgill, D.; Jobin, C. Campylobacter jejuni induces colitis through activation of mammalian target of rapamycin signaling. Gastroenterology 2012, 142, 86–95.e5.
- Konkel, M.E.; Talukdar, P.K.; Negretti, N.M.; Klappenbach, C.M. Taking Control: Campylobacter jejuni Binding to Fibronectin Sets the Stage for Cellular Adherence and Invasion. Front. Microbiol. 2020, 11, 564.
- Attene-Ramos, M.S.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. Evidence that hydrogen sulfide is a genotoxic agent. Mol. Cancer Res. 2006, 4, 9–14.
- Dahmus, J.D.; Kotler, D.L.; Kastenberg, D.M.; Kistler, C.A. The gut microbiome and colorectal cancer: A review of bacterial pathogenesis. J. Gastrointest. Oncol. 2018, 9, 769–777.
- Kapral, M.; Weglarz, L.; Parfiniewicz, B.; Lodowska, J.; Jaworska-Kik, M. Quantitative evaluation of transcriptional activation of NF-kappaB p65 and p50 subunits and IkappaBalpha encoding genes in colon cancer cells by Desulfovibrio desulfuricans endotoxin. Folia Microbiol. 2010, 55, 657–661.
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127.
- Olsen, I.; Yilmaz, O. Possible role of Porphyromonas gingivalis in orodigestive cancers. J. Oral Microbiol. 2019, 11, 1563410.
- Zhou, L.; Dorfer, C.E.; Chen, L.; Fawzy El-Sayed, K.M. Porphyromonas gingivalis lipopolysaccharides affect gingival stem/progenitor cells attributes through NF-kappaB, but not Wnt/beta-catenin, pathway. J. Clin. Periodontol. 2017, 44, 1112–1122.
- Al-Attar, A.; Alimova, Y.; Kirakodu, S.; Kozal, A.; Novak, M.J.; Stromberg, A.J.; Orraca, L.; Gonzalez-Martinez, J.; Martinez, M.; Ebersole, J.L.; et al. Activation of Notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA(2)-IIA. Mucosal Immunol. 2018, 11, 1047–1059.
- Burotto, M.; Wilkerson, J.; Stein, W.D.; Bates, S.E.; Fojo, T. Adjuvant and neoadjuvant cancer therapies: A historical review and a rational approach to understand outcomes. Semin. Oncol. 2019, 46, 83–99.
- Liedtke, C.; Mazouni, C.; Hess, K.R.; Andre, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281.
- Gonzalez-Angulo, A.M.; Morales-Vasquez, F.; Hortobagyi, G.N. Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol. 2007, 608, 1–22.
- Einhorn, L.H. Treatment of testicular cancer: A new and improved model. J. Clin. Oncol. 1990, 8, 1777–1781.
- Poletto, S.; Novo, M.; Paruzzo, L.; Frascione, P.M.M.; Vitolo, U. Treatment strategies for patients with diffuse large B-cell lymphoma. Cancer Treat. Rev. 2022, 110, 102443.
- Anand, U.; Dey, A.; Chandel, A.K.S.; Sanyal, R.; Mishra, A.; Pandey, D.K.; De Falco, V.; Upadhyay, A.; Kandimalla, R.; Chaudhary, A.; et al. Cancer chemotherapy and beyond: Current status, drug candidates, associated risks and progress in targeted therapeutics. Genes Dis. 2023, 10, 1367–1401.
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillere, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976.
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160.
- Tsai, H.F.; Hsu, P.N. Cancer immunotherapy by targeting immune checkpoints: Mechanism of T cell dysfunction in cancer immunity and new therapeutic targets. J. Biomed. Sci. 2017, 24, 35.
- Johnson, D.B.; Nebhan, C.A.; Moslehi, J.J.; Balko, J.M. Immune-checkpoint inhibitors: Long-term implications of toxicity. Nat. Rev. Clin. Oncol. 2022, 19, 254–267.
- Reckova, M.; Mladosievicova, B. An ongoing evolution of cardio-oncology with the rapid development of modern immunotherapy. Int. J. Cardiol. 2022, 347, 60–61.
- Zomborska, E.; Kasperova, S.; Slopovsky, J.; Pazderova, N.; Kasperova, B.; Penz, P.; Nyitrayova, O.; Salek, T.; Porsok, S.; Mladosievicova, B.; et al. Fatal myocarditis after the first dose of nivolumab. Klin. Onkol. 2022, 35, 486–492.
- Brahmer, J.R.; Abu-Sbeih, H.; Ascierto, P.A.; Brufsky, J.; Cappelli, L.C.; Cortazar, F.B.; Gerber, D.E.; Hamad, L.; Hansen, E.; Johnson, D.B.; et al. Society for Immunotherapy of Cancer (SITC) clinical practice guideline on immune checkpoint inhibitor-related adverse events. J. Immunother. Cancer 2021, 9, e002435.
- Lu, Y.; Yuan, X.; Wang, M.; He, Z.; Li, H.; Wang, J.; Li, Q. Gut microbiota influence immunotherapy responses: Mechanisms and therapeutic strategies. J. Hematol. Oncol. 2022, 15, 47.
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. 2021, 16, 223–249.
- He, Y.; Fu, L.; Li, Y.; Wang, W.; Gong, M.; Zhang, J.; Dong, X.; Huang, J.; Wang, Q.; Mackay, C.R.; et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. 2021, 33, 988–1000.e7.
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602.
- Ahmad, S.S.; Reinius, M.A.; Hatcher, H.M.; Ajithkumar, T.V. Anticancer chemotherapy in teenagers and young adults: Managing long term side effects. BMJ 2016, 354, i4567.
- Bajic, J.E.; Johnston, I.N.; Howarth, G.S.; Hutchinson, M.R. From the Bottom-Up: Chemotherapy and Gut-Brain Axis Dysregulation. Front. Behav. Neurosci. 2018, 12, 104.
- Parker, A.; Fonseca, S.; Carding, S.R. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes 2020, 11, 135–157.
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478.
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133.
- Kong, C.; Liang, L.; Liu, G.; Du, L.; Yang, Y.; Liu, J.; Shi, D.; Li, X.; Ma, Y. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut 2023, 72, 1129–1142.
- Janelsins, M.C.; Heckler, C.E.; Peppone, L.J.; Kamen, C.; Mustian, K.M.; Mohile, S.G.; Magnuson, A.; Kleckner, I.R.; Guido, J.J.; Young, K.L.; et al. Cognitive Complaints in Survivors of Breast Cancer After Chemotherapy Compared with Age-Matched Controls: An Analysis from a Nationwide, Multicenter, Prospective Longitudinal Study. J. Clin. Oncol. 2017, 35, 506–514.
- Chovanec, M.; Vasilkova, L.; Setteyova, L.; Obertova, J.; Palacka, P.; Rejlekova, K.; Sycova-Mila, Z.; Kalavska, K.; Svetlovska, D.; Cingelova, S.; et al. Long-Term Cognitive Functioning in Testicular Germ-Cell Tumor Survivors. Oncologist 2018, 23, 617–623.
- Lyon, A.R.; Lopez-Fernandez, T.; Couch, L.S.; Asteggiano, R.; Aznar, M.C.; Bergler-Klein, J.; Boriani, G.; Cardinale, D.; Cordoba, R.; Cosyns, B.; et al. 2022 ESC Guidelines on cardio-oncology developed in collaboration with the European Hematology Association (EHA), the European Society for Therapeutic Radiology and Oncology (ESTRO) and the International Cardio-Oncology Society (IC-OS). Eur. Heart J. 2022, 43, 4229–4361.
- Couch, L.S.; Lyon, A.R.; Lopez-Fernandez, T. Cardio-oncology: A new field requiring guidance. Eur. Heart J. Cardiovasc. Imaging 2023, 24, e47.
- Drobni, Z.D.; Alvi, R.M.; Taron, J.; Zafar, A.; Murphy, S.P.; Rambarat, P.K.; Mosarla, R.C.; Lee, C.; Zlotoff, D.A.; Raghu, V.K.; et al. Association Between Immune Checkpoint Inhibitors with Cardiovascular Events and Atherosclerotic Plaque. Circulation 2020, 142, 2299–2311.
- Mladosievicova, B.; Petrikova, L.; Valaskova, Z.; Bernadic, M., Jr.; Chovanec, M.; Mego, M.; Bernadic, M., Sr. Atherosclerosis in cancer patients. Bratisl. Lek. Listy 2019, 120, 636–640.
- Mertens, A.C.; Yasui, Y.; Neglia, J.P.; Potter, J.D.; Nesbit, M.E., Jr.; Ruccione, K.; Smithson, W.A.; Robison, L.L. Late mortality experience in five-year survivors of childhood and adolescent cancer: The Childhood Cancer Survivor Study. J. Clin. Oncol. 2001, 19, 3163–3172.
- Grilz, E.; Posch, F.; Nopp, S.; Konigsbrugge, O.; Lang, I.M.; Klimek, P.; Thurner, S.; Pabinger, I.; Ay, C. Relative risk of arterial and venous thromboembolism in persons with cancer vs. persons without cancer-a nationwide analysis. Eur. Heart J. 2021, 42, 2299–2307.
- Florido, R.; Daya, N.R.; Ndumele, C.E.; Koton, S.; Russell, S.D.; Prizment, A.; Blumenthal, R.S.; Matsushita, K.; Mok, Y.; Felix, A.S.; et al. Cardiovascular Disease Risk among Cancer Survivors: The Atherosclerosis Risk in Communities (ARIC) Study. J. Am. Coll. Cardiol. 2022, 80, 22–32.
- Herrema, H.; Nieuwdorp, M.; Groen, A.K. Microbiome and Cardiovascular Disease. In Prevention and Treatment of Atherosclerosis: Improving State-of-the-Art Management and Search for Novel Targets; von Eckardstein, A., Binder, C.J., Eds.; Springer: Cham, Switzerland, 2022; pp. 311–334.
- Zhu, W.; Wang, Z.; Tang, W.H.W.; Hazen, S.L. Gut Microbe-Generated Trimethylamine N-Oxide From Dietary Choline Is Prothrombotic in Subjects. Circulation 2017, 135, 1671–1673.
- Tang, W.H.; Hazen, S.L. The contributory role of gut microbiota in cardiovascular disease. J. Clin. Investig. 2014, 124, 4204–4211.
- Heianza, Y.; Ma, W.; Manson, J.E.; Rexrode, K.M.; Qi, L. Gut Microbiota Metabolites and Risk of Major Adverse Cardiovascular Disease Events and Death: A Systematic Review and Meta-Analysis of Prospective Studies. J. Am. Heart Assoc. 2017, 6, e004947.
- Velasquez, M.T.; Ramezani, A.; Manal, A.; Raj, D.S. Trimethylamine N-Oxide: The Good, the Bad and the Unknown. Toxins 2016, 8, 326.
- Washburn, R.L.; Cox, J.E.; Muhlestein, J.B.; May, H.T.; Carlquist, J.F.; Le, V.T.; Anderson, J.L.; Horne, B.D. Pilot Study of Novel Intermittent Fasting Effects on Metabolomic and Trimethylamine N-oxide Changes During 24-hour Water-Only Fasting in the FEELGOOD Trial. Nutrients 2019, 11, 246.
- Ciernikova, S.; Sevcikova, A.; Drgona, L.; Mego, M. Modulating the gut microbiota by probiotics, prebiotics, postbiotics, and fecal microbiota transplantation: An emerging trend in cancer patient care. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188990.
- Huang, F.; Li, S.; Chen, W.; Han, Y.; Yao, Y.; Yang, L.; Li, Q.; Xiao, Q.; Wei, J.; Liu, Z.; et al. Postoperative Probiotics Administration Attenuates Gastrointestinal Complications and Gut Microbiota Dysbiosis Caused by Chemotherapy in Colorectal Cancer Patients. Nutrients 2023, 15, 356.
- Mego, M.; Chovanec, J.; Vochyanova-Andrezalova, I.; Konkolovsky, P.; Mikulova, M.; Reckova, M.; Miskovska, V.; Bystricky, B.; Beniak, J.; Medvecova, L.; et al. Prevention of irinotecan induced diarrhea by probiotics: A randomized double blind, placebo controlled pilot study. Complement. Ther. Med. 2015, 23, 356–362.
- Mego, M.; Danis, R.; Chovanec, J.; Jurisova, S.; Bystricky, B.; Porsok, S.; Konkolovsky, P.; Vaclav, V.; Wagnerova, M.; Stresko, M.; et al. Randomized double-blind, placebo-controlled multicenter phase III study of prevention of irinotecan-induced diarrhea by a probiotic mixture containing Bifidobacterium BB-12((R))Lactobacillus rhamnosus LGG((R)) in colorectal cancer patients. Front. Oncol. 2023, 13, 1168654.
- Cui, M.; Xiao, H.; Li, Y.; Zhou, L.; Zhao, S.; Luo, D.; Zheng, Q.; Dong, J.; Zhao, Y.; Zhang, X.; et al. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol. Med. 2017, 9, 448–461.
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103.
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97.
- Tintelnot, J.; Xu, Y.; Lesker, T.R.; Schonlein, M.; Konczalla, L.; Giannou, A.D.; Pelczar, P.; Kylies, D.; Puelles, V.G.; Bielecka, A.A.; et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 2023, 615, 168–174.
