Forests represent a vital natural resource and play a crucial role in climate regulation and maintaining biodiversity. However, the growth and development of forest trees are increasingly challenged by rising environmental pressures, particularly detrimental abiotic stressors. To address these challenges, genetic transformation technologies have emerged as effective solutions. Despite various difficulties in genetic transformation for forest trees, including prolonged life cycles, genetic diversity, interspecies variations, and complex regeneration systems, significant research progress has been achieved in tree gene editing, transgenic technology, and methods for delivering exogenous molecules. These technologies have the potential to enhance tree quality, increase productivity, and improve resistance to abiotic stress. This review provides an overview of the main methods and transformation receptors in tree genetic transformation. Additionally, we summarize several novel techniques, such as nanoparticle-mediated gene transformation, advanced gene editing technology, various novel delivery carriers, and non-genetically modified protein function interference through peptide aptamer. Notably, we also place emphasis on several referable genes from forest trees and common crops, together with their potential function for improving abiotic stress responses. Through this research, we aspire to achieve sustainable utilization and conservation of tree resources, thereby providing substantial support for future livelihoods and economic development.
- genetic transformation
- transformation receptor
- abiotic stress
- nanoparticle
- peptide aptamer
1. Introduction
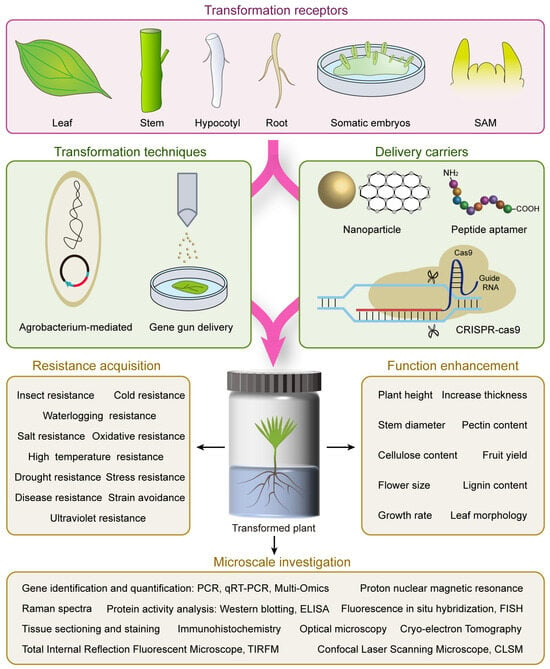
2. Tree Genetic Transformation Techniques and Transformation Receptor
2.1. Common Tree Genetic Transformation Techniques
2.2. Transformation Receptor Used in Forest Tree Genetic Transformation
3. Advancements in Tree Genetic Transformation Process
3.1. Nanoparticle-Meditated Gene Transformation
3.2. Optimized Gene Editing Technologies
3.3. DNA Free Gene Editing Technology
3.4. Peptide Aptamer Meditated Non-Genetically Modified for Protein Function Interference
References
- Liang, J.; Crowther, T.W.; Picard, N.; Wiser, S.; Zhou, M.; Alberti, G.; Schulze, E.D.; McGuire, A.D.; Bozzato, F.; Pretzsch, H.; et al. Positive biodiversity-productivity relationship predominant in global forests. Science 2016, 354, aaf8957.
- José, A.-S.; Luis, B.U.A.; María, L.-S.; Juan, V.M.O. Forest ecosystem services: An analysis of worldwide research. Forests 2018, 9, 453.
- Suzuki, S.; Suzuki, H. Recent advances in forest tree biotechnology. Plant Biotechnol. 2014, 31, 1–9.
- Thapliyal, G.; Bhandari, M.S.; Vemanna, R.S.; Pandey, S.; Meena, R.K.; Barthwal, S. Engineering traits through CRISPR/cas genome editing in woody species to improve forest diversity and yield. Crit. Rev. Biotechnol. 2022, 43, 884–903.
- Forner, J.; Kleinschmidt, D.; Meyer, E.H.; Gremmels, J.; Morbitzer, R.; Lahaye, T.; Schottler, M.A.; Bock, R. Targeted knockout of a conserved plant mitochondrial gene by genome editing. Nat. Plants 2023, 9, 1818–1831.
- Cardi, T. Cisgenesis and genome editing: Combining concepts and efforts for a smarter use of genetic resources in crop breeding. Plant Breed. 2016, 135, 139–147.
- Jo, K.R.; Kim, C.J.; Kim, S.J.; Kim, T.Y.; Bergervoet, M.; Jongsma, M.A.; Visser, R.G.; Jacobsen, E.; Vossen, J.H. Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol. 2014, 14, 50.
- Wingfield, M.J.; Brockerhoff, E.G.; Wingfield, B.D.; Slippers, B. Planted forest health: The need for a global strategy. Science 2015, 349, 832–836.
- Yin, Y.; Wang, C.; Xiao, D.; Liang, Y.; Wang, Y. Advances and perspectives of transgenic technology and biotechnological application in forest trees. Front. Plant Sci. 2021, 12, 786328.
- De Meester, B.; Vanholme, R.; Mota, T.; Boerjan, W. Lignin engineering in forest trees: From gene discovery to field trials. Plant Commun. 2022, 3, 100465.
- De Cleene, M.; De Ley, J. The host range of crown gall. Bot. Rev. 1976, 42, 389–466.
- Sanford, J.C.; Klein, T.M.; Wolf, E.D.; Allen, N. Delivery of substances into cells and tissues using a particle bombardment process. Part. Sci. Technol. 1987, 5, 27–37.
- Ikeuchi, M.; Sugimoto, K.; Iwase, A. Plant callus: Mechanisms of induction and repression. Plant Cell 2013, 25, 3159–3173.
- Nagle, M.; Dejardin, A.; Pilate, G.; Strauss, S.H. Opportunities for innovation in genetic transformation of forest trees. Front. Plant Sci. 2018, 9, 1443.
- Sharma, V.; Magotra, T.; Chourasia, A.; Mittal, D.; Singh, U.P.; Sharma, S.; Sharma, S.; Ramirez, Y.G.; Dobranszki, J.; Martinez-Montero, M.E. Thin cell layer tissue culture technology with emphasis on tree species. Forests 2023, 14, 1212.
- Arokiaraj, P.; Jones, H.; Cheong, K.F.; Coomber, S.; Charlwood, B.V. Gene insertion into Hevea brasiliensis. Plant Cell Rep. 1994, 13, 425–431.
- Tang, W.; Tian, Y. Transgenic loblolly pine (Pinus taeda L.) plants expressing a modified δ-endotoxin gene of Bacillus thuringiensis with enhanced resistance to Dendrolimus punctatus Walker and Crypyothelea formosicola Staud. J. Exp. Bot. 2003, 54, 835–844.
- Grant, J.E.; Cooper, P.A.; Dale, T.M. Genetic transformation of micropropagated shoots of Pinus radiata D.Don. bioRxiv 2015.
- Lee, H.; Moon, H.-K.; Park, S.-Y. Agrobacterium-mediated transformation via somatic embryogenesis system in korean fir (Abies koreana Wil.), a korean native conifer. Korean J. Plant Resour. 2014, 27, 242–248.
- Tang, W.; Xiao, B.; Fei, Y. Slash pine genetic transformation through embryo cocultivation with A. tumefaciens and transgenic plant regeneration. In Vitro Cell. Dev. Biol.-Plant 2013, 50, 199–209.
- Montoya, A.L.; Chilton, M.D.; Gordon, M.P.; Sciaky, D.; Nester, E.W. Octopine and nopaline metabolism in Agrobacterium tumefaciens and crown gall tumor cells: Role of plasmid genes. J. Bacteriol. 1977, 129, 101–107.
- Song, Y.; Bai, X.; Dong, S.; Yang, Y.; Dong, H.; Wang, N.; Zhang, H.; Li, S. Stable and sfficient Agrobacterium-mediated genetic transformation of larch using embryogenic callus. Front. Plant Sci. 2020, 11, 584492.
- Horsch, R.B.; Klee, H.J. Rapid assay of foreign gene expression in leaf discs transformed by Agrobacterium tumefaciens: Role of T-DNA borders in the transfer process. Proc. Natl. Acad. Sci. USA 1986, 83, 4428–4432.
- Xia, Y.; Cao, Y.; Ren, Y.; Ling, A.; Du, K.; Li, Y.; Yang, J.; Kang, X. Effect of a suitable treatment period on the genetic transformation efficiency of the plant leaf disc method. Plant Methods 2023, 19, 15.
- Samaneh Sadat, M. Study on factors influencing transformation efficiency in Pinus massoniana using Agrobacterium tumefaciens. Plant Cell Tiss. Org. 2018, 133, 437–445.
- Taniguchi, T.; Ohmiya, Y.; Kurita, M.; Tsubomura, M.; Kondo, T. Regeneration of transgenic Cryptomeria japonica D. Don after Agrobacterium tumefaciens-mediated transformation of embryogenic tissue. Plant Cell Rep. 2008, 27, 1461–1466.
- Bruegmann, T.; Wetzel, H.; Hettrich, K.; Smeds, A.; Willför, S.; Kersten, B.; Fladung, M. Knockdown of PCBER1, a gene of neolignan biosynthesis, resulted in increased poplar growth. Planta 2018, 249, 515–525.
- Tulecke, W.; McGranahan, G.; Ahmadi, H. Regeneration by somatic embryogenesis of triploid plants from endosperm of walnut, Juglans regia L. cv Manregian. Plant Cell Rep. 1988, 7, 301–304.
- Verdeil, J.-L.; Alemanno, L.; Niemenak, N.; Tranbarger, T.J. Pluripotent versus totipotent plant stem cells: Dependence versus autonomy? Trends Plant Sci. 2007, 12, 245–252.
- Hu, R.; Sun, Y.; Wu, B.; Duan, H.; Zheng, H.; Hu, D.; Lin, H.; Tong, Z.; Xu, J.; Li, Y. Somatic embryogenesis of immature Cunninghamia lanceolata (Lamb.) Hook zygotic embryos. Sci. Rep. 2017, 7, 56.
- San José, M.C.; Cernadas, M.J.; Corredoira, E. Histology of the regeneration of Paulownia tomentosa (Paulowniaceae) by organogenesis. Rev. Biol. Trop. 2014, 62, 809–818.
- Cao, X.; Xie, H.; Song, M.; Lu, J.; Ma, P.; Huang, B.; Wang, M.; Tian, Y.; Chen, F.; Peng, J.; et al. Cut–dip–budding delivery system enables genetic modifications in plants without tissue culture. Innovation 2023, 4, 100345.
- Meng, D.; Yang, Q.; Dong, B.; Song, Z.; Niu, L.; Wang, L.; Cao, H.; Li, H.; Fu, Y. Development of an efficient root transgenic system for pigeon pea and its application to other important economically plants. Plant Biotechnol. J. 2019, 17, 1804–1813.
- López, M.; Humara, J.M.; Rodríguez, R.; Ordás, R.J. Transient uidA gene expression in electroporated cotyledonary protoplasts of Pinus nigra ssp. salzmannii and in bombarded cotyledons. Can. J. For. Res. 2000, 30, 448–455.
- Salaj, T.; Klubicová, K.; Matusova, R.; Salaj, J. Somatic embryogenesis in selected conifer trees Pinus nigra Arn. and Abies hybrids. Front. Plant Sci. 2019, 10, 13.
- Zhu, T.; Moschou, P.N.; Alvarez, J.M.; Sohlberg, J.J.; von Arnold, S. Wuschel-related homeobox 8/9 is important for proper embryo patterning in the gymnosperm Norway spruce. J. Exp. Bot. 2014, 65, 6543–6552.
- Mei, G.; Chen, A.; Wang, Y.; Li, S.; Wu, M.; Hu, Y.; Liu, X.; Hou, X. A simple and efficient in planta transformation method based on the active regeneration capacity of plants. Plant Commun. 2024, 2024, 100822.
- Anders, C.; Hoengenaert, L.; Boerjan, W. Accelerating wood domestication in forest trees through genome editing: Advances and prospects. Curr. Opin. Plant Biol. 2023, 71, 102329.
- Yao, W.; Zhao, K.; Cheng, Z.; Li, X.; Zhou, B.; Jiang, T. Transcriptome analysis of poplar under salt stress and over-expression of transcription factor NAC57 geneconfers salt tolerance in transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1121.
- Wang, P.; Lombi, E.; Zhao, F.J.; Kopittke, P.M. Nanotechnology: A new opportunity in plant sciences. Trends Plant Sci. 2016, 21, 699–712.
- Peng, L.H.; Gu, T.W.; Xu, Y.; Dad, H.A.; Liu, J.X.; Lian, J.Z.; Huang, L.Q. Gene delivery strategies for therapeutic proteins production in plants: Emerging opportunities and challenges. Biotechnol. Adv. 2022, 54, 107845.
- Li, Y.; Bao, W.; Wu, H.; Wang, J.; Zhang, Y.; Wan, Y.; Cao, D.; O’Hare, D.; Wang, Q. Delaminated layered double hydroxide delivers DNA molecules as sandwich nanostructure into cells via a non-endocytic pathway. Sci. Bull. 2017, 62, 686–692.
- Su, W.B.; Xu, M.Y.; Radani, Y.; Yang, L.M. Technological development and application of plant genetic transformation. Int. J. Mol. Sci. 2023, 24, 10646.
- Bewg, W.P.; Ci, D.; Tsai, C.J. Genome editing in trees: From multiple repair pathways to long-term stability. Front. Plant Sci. 2018, 9, 1732.
- Miladinovic, D.; Antunes, D.; Yildirim, K.; Bakhsh, A.; Cvejić, S.; Kondić-Špika, A.; Marjanovic Jeromela, A.; Opsahl-Sorteberg, H.-G.; Zambounis, A.; Hilioti, Z. Targeted plant improvement through genome editing: From laboratory to field. Plant Cell Rep. 2021, 40, 935–951.
- Flachowsky, H.; Peil, A.; Sopanen, T.; Elo, A.; Hanke, V. Overexpression of BpMADS4 from silver birch (Betula pendula Roth.) induces early-flowering in apple (Malus × domestica Borkh.). Plant Breed. 2007, 126, 137–145.
- Cardi, T.; Murovec, J.; Bakhsh, A.; Boniecka, J.; Bruegmann, T.; Bull, S.E.; Eeckhaut, T.; Fladung, M.; Galovic, V.; Linkiewicz, A.; et al. CRISPR/Cas-mediated plant genome editing: Outstanding challenges a decade after implementation. Trends Plant Sci. 2023, 28, 1144–1165.
- Xue, C.; Qiu, F.; Wang, Y.; Li, B.; Zhao, K.T.; Chen, K.; Gao, C. Tuning plant phenotypes by precise, graded downregulation of gene expression. Nat. Biotechnol. 2023, 41, 1758–1764.
- Huang, J.; Lin, Q.; Fei, H.; He, Z.; Xu, H.; Li, Y.; Qu, K.; Han, P.; Gao, Q.; Li, B.; et al. Discovery of deaminase functions by structure-based protein clustering. Cell 2023, 186, 3182–3195.
- Kocsisova, Z.; Coneva, V. Strategies for delivery of CRISPR/Cas-mediated genome editing to obtain edited plants directly without transgene integration. Front. Genome Ed. 2023, 5, 1209586.
- Khosravi, S.; Ishii, T.; Dreissig, S.; Houben, A. Application and prospects of CRISPR/Cas9-based methods to trace defined genomic sequences in living and fixed plant cells. Chromosome Res. 2020, 28, 7–17.
- He, Y.; Zhu, M.; Wang, L.; Wu, J.; Wang, Q.; Wang, R.; Zhao, Y. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant. 2018, 11, 1210–1213.
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617.
- Ma, X.; Li, X.; Li, Z. Transgene-free genome editing in Nicotiana benthamiana with CRISPR/Cas9 delivered by a rhabdovirus vector. In Plant Genome Engineering: Nethods and Protocols; Yang, B., Harwood, W., Que, Q., Eds.; Springer US: New York, NY, USA, 2023; Volume 2653, pp. 173–185.
- Hao, Z.; Gong, P.; He, C.; Lin, J. Peptide aptamers to inhibit protein function in plants. Trends Plant Sci. 2018, 23, 281–284.
- Gong, P.; Quan, H.; He, C. Targeting MAGO proteins with a peptide aptamer reinforces their essential roles in multiple rice developmental pathways. Plant J. 2014, 80, 905–914.
- Torti, S.; Schlesier, R.; Thummler, A.; Bartels, D.; Romer, P.; Koch, B.; Werner, S.; Panwar, V.; Kanyuka, K.; Wiren, N.V.; et al. Transient reprogramming of crop plants for agronomic performance. Nat. Plants 2021, 7, 159–171.
- Colombo, M.; Mizzotti, C.; Masiero, S.; Kater, M.M.; Pesaresi, P. Peptide aptamers: The versatile role of specific protein function inhibitors in plant biotechnology. J. Integr. Plant Biol. 2015, 57, 892–901.
- Ormancey, M.; Guillotin, B.; Merret, R.; Camborde, L.; Duboé, C.; Fabre, B.; Pouzet, C.; Impens, F.; Van Haver, D.; Carpentier, M.-C.; et al. Complementary peptides represent a credible alternative to agrochemicals by activating translation of targeted proteins. Nat. Commun. 2023, 14, 254.
