The intracellular orchestra of protein synthesis unfolds through a delicate interplay of small nucleolar RNAs (snoRNAs), small nucleolar RNA host genes (SNHGs), ribosomal RNAs (rRNAs), the nucleolus, and ribosomes. snoRNAs, derived from specific genomic loci, often SNHGs, intricately direct the modification and processing of rRNAs within the nucleolus. Mature rRNAs form the architectural backbone of ribosomes, the molecular machines that translate genetic information into functional proteins. In recent years, snoRNAs and their associated factors have attracted growing attention. Numerous publications have revealed that snoRNAs are implicated in human diseases, particularly cancer, to a greater extent than previously believed.
2. Characteristics and General Functions of snoRNA
2.1. Classification of snoRNAs
2.1.1. C/D Box
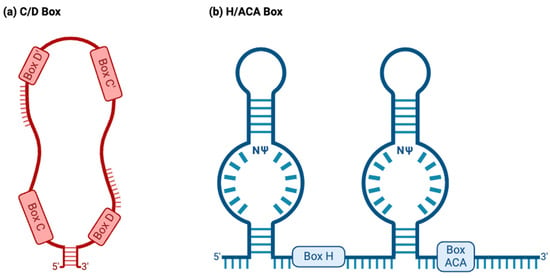
C/D box snoRNAs, collectively known as SNORDs, are characterized by their kink-turn (also called the stem–bulge–stem) structure
[8]. As their name suggests, C/D box snoRNAs contain C and D boxes, which are conserved sequence motifs present in canonical C/D box snoRNAs (
Figure 2a)
[43]. C boxes consist of the sequence motif RUGAUGA (R standing in for a purine base), while D boxes represent the sequence CUGA
[44]. In addition, C/D box snoRNAs contain a less conserved C′ and D′ box located upstream of the D box and downstream of the C box, respectively (
Figure 2a), which have identical nucleotides to their counterparts
[45]. Upstream of the D and D′ boxes are antisense elements known as “guide regions”, which are complementary to target RNAs and allow for the precise methylation of targets (
Figure 2a)
[44]. Using their scaffold-like structure, conventional SNORDS attract and position partner proteins, canonically including the (human/yeast) fibrillarin (FBL)/Nop1p, SNU13 (NHP2L1)/Snu13p, NOP58/Nop58p, and NOP56/Nop56p to form a small nucleolar ribonucleoprotein (snoRNP) (
Figure 3a)
[44][46][44,46]. Partner proteins appeared to interact reciprocally in the accumulation of snoRNAs as well
[47]. snoRNP complexes facilitate the canonical function of SNORDs: transferring a methyl group to the 2′ oxygen on a ribose molecule in a process known as 2′-O-methylation (
Figure 3a)
[44]. These proteins give the snoRNP functionality and prevent its exonuclease-mediated degradation while ensuring proper localization within the nucleolus
[15]. Some C/D box snoRNAs were found to facilitate the acetylation of the critical 18S structural rRNA in eukaryotes, allowing acetyltransferases to catalyze the formation of the ac4C modification
[43].
Figure 2. Two major snoRNA families. Typically, snoRNAs are classified into the C/D box or H/ACA box families based on their structure and function. (a) C/D box snoRNAs normally have a stem–bulge–stem structure, with C and D box motifs and C′ and D′ box motifs present on opposing ends of the C/D box snoRNA. The C and D boxes, located at the 5′ and 3′ ends, represent the sequence motifs RUGAUGA (R represents a purine base) and CUGA, respectively. C/D box snoRNAs canonically guide the 2′O-methylation of target RNAs, binding to targets using antisense elements located downstream of the D and D′ box sequence motifs (depicted above). (b) H/ACA box snoRNAs normally have a hairpin–hinge–hairpin–tail structure, with the H box present on the hinge and the ACA box located on the tail. The H and ACA boxes represent the motifs ANANNA and ACA. H/ACA box snoRNAs also possess complimentary guide sequences. However, these appear within “pseudouridylation pockets”, in which the modification occurs. NΨ: symbol for a pseudouridine nucleotide, representing the location on H/ACA box snoRNAs where uridines are pseudouridylated.
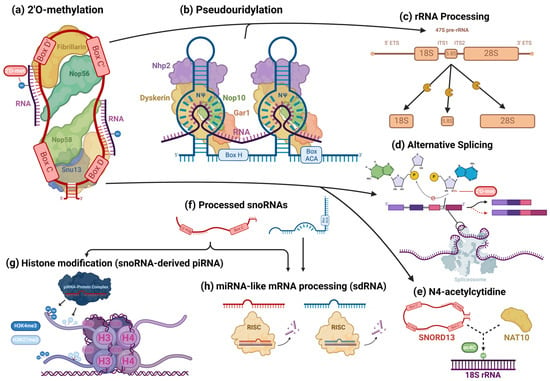
Figure 3. Cellular processes facilitated by snoRNAs. snoRNAs are mostly associated with their role as guides for 2′O-methylation (a) and pseudouridylation (b) modifications, in which they bind to target transcripts and, using their scaffold-like structure, coordinate snoRNP core proteins that catalyze these modifications. However, snoRNAs are also involved in the epigenetic regulation of a wide variety of targets. In addition to their roles in rRNA processing (c), snoRNAs also regulate alternative splicing (d) and can guide rarer nucleotide modifications, such as the N4-acetylcytidine modification (e). Some snoRNAs are processed into smaller fragments (f) and form complexes with piRNA- and miRNA-associated proteins, playing the role of those RNAs. snoRNA-derived piRNAs and miRNAs have been found to regulate histone modifications (g) and cleave target RNAs (h), respectively, modulating gene expression.
2.1.2. H/ACA Box
H/ACA box snoRNAs, known as SNORAs, also contain the kink-turn structure. However, they contain two additional hairpin structures and different sequence motifs and have corresponding differences in functions (
Figure 2b)
[43]. H/ACA box snoRNAs’ canonical function is to facilitate the RNA-dependent pseudouridylation of their target RNAs, which primarily consist of pre-rRNAs
[48]. Pseudouridine is the most common isomer of uridine, and the pseudouridylation of rRNAs is critical for proper ribosomal function in eukaryotes
[48]. These RNAs’ hinge (H) and ACA boxes, which consist of the sequence motifs ANANNA and ACA, respectively, are located downstream of their kink-turn structures (
Figure 2b)
[44]. Within the internal loops of an H/ACA box snoRNA are pseudouridylation pockets containing antisense elements, which bind to target RNA via complementary base pairing and allow for the precise pseudouridylation of targets (
Figure 2b)
[48]. Conventional H/ACA snoRNAs form snoRNP complexes with NHP2/Nhp2p, NOP10/Nop10p, GAR1/Gar1p, and the pseudouridine synthase dyskerin (DKC1)/Cbf5p (
Figure 3b)
[44][46][44,46].
2.1.3. scaRNAs
Lastly, the small Cajal body RNA (scaRNA) subfamily consists of snoRNAs with the structural characteristics of the C/D box and H/ACA RNAs and occasionally both
[43]. Although scaRNAs can fall under the C/D-H/ACA classification, their unique localization in nuclear Cajal bodies distinguishes them from other families. scaRNAs also contain distinct sequence motifs, including the CAB box and GU-repeat elements, which are necessary for the proper localization of scaRNAs in Cajal bodies
[49]. Within Cajal bodies, scaRNAs guide 2′-O-methylation, pseudouridylation, and other modifications to process various RNA species, similarly to their C/D and H/ACA box counterparts
[44].
2.2. Canonical snoRNA/snoRNP Biogenesis
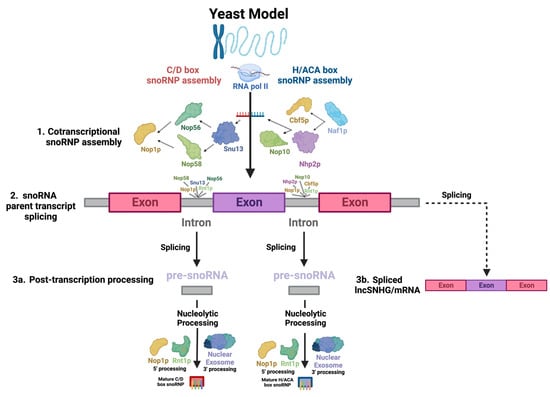
The common pathways of snoRNA biogenesis are well established in literature. As the majority of studies that characterize this elaborate process were conducted on yeast samples, the processes that will be discussed occurred in yeast unless otherwise specified, and the yeast homologs of proteins will be named, though the process is assumed to be comparable to that of humans due to the high degree of conservation between human and yeast intron-derived snoRNAs
[15][50][15,50]. Still, the use of this model species presents limitations to the current understandings of snoRNA biogenesis in humans, which additional studies are needed to clarify.
Most human snoRNAs are embedded in the introns of noncoding snoRNA host genes (SNHGs). However, some snoRNAs are found within the introns of protein-coding genes (
Figure 1(2): snoRNA parent transcript splicing)
[51]. snoRNAs can be transcribed in a mono- or polycistronic manner, though polycistronic clustering occurs infrequently in humans
[51]. Human snoRNA-embedded primary transcripts are transcribed by RNA polymerase II
[51]. Nop1p binds to snoRNAs and associates with Rnt1p, an endonuclease which binds to the external stem structural motif. Rnt1p and Nop1p then cooperatively cleave snoRNAs confined in mono- and polycistronic sequences from the primary transcript into intron lariats, which are enzymatically debranched and linearized and trimmed by exoribonucleases packaged in the nuclear exosome in order to become functional transcripts (
Figure 1(2): snoRNA parent transcript splicing)
[52][53][54][55][52,53,54,55]. snoRNAs undergo slightly different processes at the 5′ and 3′ ends, with Nop1p and Rnt1p targeting sites present on the 5′ end and removing the 5′ cap, while on the 3′ end, endonucleolytic cleavages map entry points for exonucleases to continue processing the snoRNA (
Figure 1(3a): Post-transcription processing)
[52][56][52,56].
Simultaneously, to protect the molecule from immediate post-transcriptional exonucleolytic degradation, snoRNP proteins assemble cotranscriptionally on nascent snoRNA transcripts, forming precursors to the canonical snoRNP
[57][58][57,58]. For H/ACA snoRNAs, Naf1p, a H/ACA snoRNP assembly factor, recruits Cbf5p and Nhp2p, the catalytic subunit of H/ACA snoRNPs—the yeast homolog of dyskerin
[59]—and a H/ACA snoRNP core protein, respectively, which associate with the C-terminus of the RNA polymerase II (RPB1) subunit
[58] at the 3′ end of the snoRNA
[59], which recruits several other RNA-binding proteins, including Nop10, a canonical snoRNP core protein (
Figure 1(1): Cotranscriptional snoRNP assembly)
[57][59][57,59]. Gar1p is posttranscriptionally recruited, indicating that it is not directly involved in snoRNA synthesis
[57]. SHQ1 may also play a vital role as a chaperone molecule for Cbf5p
[60]. For human C/D box snoRNAs, the mature snoRNP complex is sequentially assembled, beginning with Snu13, which recognizes structural motifs of the C/D and C′/D′ boxes, forming a precursor complex which is recognized by Nop56 and Nop58, whose N-termini recruit Fibrillarin (
Figure 1(1): Cotranscriptional snoRNP assembly)
[58]. Both assembly pathways likely largely involve the Hsp90 and R2TP complex chaperone molecules, which stabilize RNP core proteins and coordinate a large array of assembly factors
[61].
snoRNAs can generally possess two different kinds of 5′ cap modifications, including the 7-methylguanosine cap and monomethylphosphate cap
[62]. RNA polymerase II-transcribed molecules generally undergo trimethylation
[62]. However, the intron-derived snoRNAs most common in humans tend to receive no cap modifications
[63]. These canonical snoRNAs lack the traditional structural features, such as the 5′ monomethylguanosine cap and poly(A) tail, present on mRNAs that undergo nuclear export
[64], which was used to explain their localization in the nucleus, though recent reports of noncanonical snoRNA activity in extranuclear cellular localizations easily disprove this notion
[64]. snoRNAs may escape the nucleus via stress-regulated transport/shuttling proteins, including NXF3 and DBR1
[55][65][55,65].
One peculiar feature of snoRNA expression—its apparent disconnection with its host gene’s expression—may be explained based on the mechanisms by which snoRNA processing occurs
[66][67][68][66,67,68]. snoRNA transcription is duly initiated with promoters containing canonical pyrimidine-anchored transcription start sites in proximal or overlapping configurations with noncanonical 5′TOP initiators to form a hybridized dual promoter
[69]. In addition, endonucleolytic cleavage serves to excise snoRNAs from their host sequences but also uncouples the expression of the two, likely as a result of nonsense-mediated decay
[70]. Thus, the unrelated expression of snoRNAs and their host genes is speculated to be a byproduct of the normal processes of snoRNA synthesis.
2.3. snoRNAs’ Canonical Roles in rRNA Processing
snoRNAs are known to play a critical role in RNA processing, primarily targeting ribosomal (rRNAs) and small nuclear RNAs (snRNAs)
[71]. In spliceosomes’ snRNAs, modified bases are concentrated in regions associated with pre-mRNA splicing and are functionally critical, regulating splice-site recognition and the formation of small nuclear ribonucleoprotein (snRNP) complexes that form the spliceosome, among other characteristics that are necessary for proper splicing
[72]. Since many snoRNAs associated with cancer have some canonical interaction with rRNAs,
rwe
searchers will highlight this aspect of snoRNAs’ canonical activity. To briefly summarize the complicated involvement of snoRNAs in rRNA processing, in human nucleoli, a cohort of snoRNAs are involved, with many ribosomal proteins and pre-ribosomal factors, in the endonucleolytic cleavage of pre-rRNA transcripts surrounding internal transcribed spacers (ITS) during the initial processing of the full-length 47S pre-rRNA precursor transcript to isolate ribosomal subunit pre-rRNAs for further processing (
Figure 3c)
[73]. Using their antisense complementarity, these snoRNAs, assembled into snoRNPs, base pair with pre-rRNAs at docking sites throughout the precursor transcript and participate in the regulation of the transcript’s structure
[73]. During this stage, snoRNPs can prevent premature folding at their binding sites, while other snoRNPs concomitantly induce modifications to trigger structural changes along the precursor transcript; the snoRNPs simultaneously prevent premature misfolding at those sites and alter the pre-rRNA’s structure at other locations, while endonucleases facilitate the cleavage of the precursor transcript, ultimately ensuring the excision of structurally correct pre-rRNA transcripts
[73][74][75][76][77][78][73,74,75,76,77,78]. Once this process is completed, RNA helicases and other RNA-binding proteins trigger the release of the snoRNPs
[73][78][73,78]. snoRNPs mediate modifications, which alter functional groups of nucleotides and, consequently, intramolecular interactions within the precursor transcript that are putatively responsible for its three-dimensional folding
[79]. However, the fact that some snoRNPs specifically bind to pre-rRNA transcripts but do not mediate any modification and are necessary for the production of functional pre-rRNA transcripts points to the existence of snoRNPs that instead direct nucleases to sites for cleavage
[80]. pre-rRNAs also undergo exonucleolytic trimming to remove external transcribed spacers via complex sets of processes, which are different for each type of pre-rRNA being processed
[73][77][73,77]. For the processing of 18S pre-rRNA, several snoRNAs and the spliceosomal subunit processome — comprised of many subcomplexes, including U3 snoRNP, which is necessary for processome formation — are essential for this trimming
[74][77][74,77].
2.4. Noncanonical snoRNA Functions
snoRNAs are a versatile RNA species involved in a range of cellular processes. Recent research has largely identified noncanonical functions for snoRNAs, which define a vast range of unexplored snoRNA activity.
Some snoRNA transcripts are further processed (
Figure 3f) by the microprocessor complex and endoribonucleases, including DICER or SLICER, into snoRNA-derived fragments (sdRNA), which are recruited into post-transcriptional silencing complexes and function similarly to miRNA (
Figure 3h)
[11][81][82][11,81,82]. Both H/ACA snoRNA and C/D snoRNA can produce miRNAs or sdRNAs. Products derived from H/ACA snoRNAs are mostly 20–24 nt long and originate from the 3′ end, while those from C/D snoRNAs exhibit a bimodal size distribution at around 17–19 nt and >27 nt, predominantly originating from the 5′ end
[12]. Similar to conventional miRNAs, many snoRNA-derived miRNAs and sdRNAs target mRNA at the 3′UTR and some at the 5′UTR or coding regions
[11]. “Orphan” snoRNAs, which show no specific RNA base-pairing complementarity, are speculated to comprise many sdRNAs
[16]. Several snoRNA-derived piRNAs have been observed to be associated with PIWI proteins and participate in epigenetic regulation in mammalian somatic cells, directing the exchange of the H3K4me3 and H3K27me3 histone modifications on gene promoter regions, binding to mRNAs, and recruiting TRAMP protein complexes, which are involved in nucleolytic RNA degradation (
Figure 3g)
[14][83][14,83]. A few C/D box snoRNAs have also been identified to participate in pre-mRNA alternative splicing. The snoRNA-mediated 2′O methylation of an intron substrate’s branch point adenosine blocks the critical transesterification reaction between its 2′ hydroxyl group and the phosphodiester bond of the guanosine and neighboring base at the intron’s 5′ splice site (
Figure 3d)
[84]. When splicing normally occurs, the phosphodiester bond is broken, resulting in a new bond between the adenosine and guanosine
[84]. The new bond forms the loop structure of the intron lariat and is necessary for proper splicing and the mature mRNA transcript’s appropriate expression
[84][85][84,85]. However, when the adenosine 2′ hydroxyl is blocked by a methyl group, exon inclusion and exclusion can occur, leading to the synthesis of alternate proteins from the same transcript (
Figure 3d). SNORD13 was found to be associated with the NAT10 acetyltransferase and guide the transfer of the N4-acetylcytidine (ac4C) modification to 18S rRNA in humans by complementary base pairing—in a manner identical to its expected canonical function (
Figure 3e)
[86]. AluACA snoRNAs, which possess an altered version of the double hairpin structure of H/ACA snoRNAs, also lack functional binding sites with canonical snoRNP proteins and are not functionally well characterized
[87]. Most noncanonical snoRNAs covered in the recent literature, however, are snoRNAs that fall under normal C/D and H/ACA box classifications and canonically guide the modification of RNAs in addition to non-canonically mediating oncogenesis. Thus, these transcripts will be the focus of this
trevie
xtw.
2.5. Small Nucleolar RNA Host Gene (SNHG)
The SNHGs are a group of genes that can be processed into snoRNAs and lncSNHGs
[9][18][9,18]. The HUGO Gene Nomenclature Committee has published more than 30 SNHGs, including
SNHG1 to
SNHG33,
GAS5, and
ZFAS1 [88]. These SNHGs are first transcribed into primary SNHG transcripts and then further spliced into exons and introns (
Figure 1)
[20][21][22][23][20,21,22,23]. The exons are then re-spliced and translocated to the cytoplasm to function as protein-coding mRNA or noncoding RNA, i.e., lncSHNGs. Intronic sequences are further processed into mature snoRNA
[9][18][9,18].
LncSNHGs are a type of lncRNA. Recently, they have attracted attention as emerging transcription regulators that function as oncogenes and tumor suppressors
[2][18][20][23][24][25][26][27][2,18,20,23,24,25,26,27]. Their regulatory roles in tumorigenesis involve various aspects of biogenesis, such as acting as miRNA sponges, inhibiting protein ubiquitination, and enhancing DNA methylation
[9][18][89][9,18,89]. Their aberrant expression also influences the abnormal behavior of cancer cells, including epithelial–mesenchymal transitions (EMT), cell cycle progression, proliferation, invasion, and the evasion of apoptosis
[28][90][91][92][93][94][28,90,91,92,93,94].
Overall, SNHGs and their lncSNHGs may play crucial roles in the future of cancer therapy. Consequently, elucidating the molecular mechanisms underlying the correlational links between these RNAs, cancer, and immune responses is necessary to contribute to the development of novel therapeutic approaches.
2.6. Nucleolus
The nucleolus, a vital membraneless organelle within the nucleus, is a central hub for rRNA processing and ribosome biogenesis
[32][33][32,33]. Dysregulated nucleolar function leads to abnormalities in ribosomal biogenesis, resulting in various ribosomopathies, including cancer
[95][96][97][95,96,97]. Structurally, the nucleolus consists of three discrete regions, namely, the fibrillar center (FC), dense fibrillar component (DFC), and granular component (GC)
[98]. Recent advancements in high-resolution live-cell microscopy have allowed for the identification of 12 proteins, including unhealthy ribosome biogenesis 1 (URB1), enriched toward the periphery of the DFC, named the peripheral dense fibrillar compartment (PDFC)
[99]. URB1, a static nucleolar protein, is crucial in anchoring and folding the 3′ end of pre-rRNA. This ensures the recognition of U8 small nucleolar RNA and the subsequent removal of the 3′ external transcribed spacer (ETS). The depletion of URB1 disrupts the DFC, causing aberrant pre-rRNA movement and altered conformation, activating exosome-dependent nucleolar surveillance with downstream effects on rRNA production and embryonic development.
Another nucleolar protein, polyglutamine-binding protein 5 (PQBP5 or NOL10), binds to polyglutamine tract sequences and constitutes the skeletal structure of the nucleolus
[100]. This protein remains stable under stress conditions, anchoring other nucleolar proteins during osmotic stress and maintaining the nucleolar structure. The functional depletion of PQBP5/NOL10, as seen in polyglutamine disease proteins, leads to pathological nucleolar deformities or disappearance.
Another notable development in the study of the nucleolus was made in the field of liquid–liquid phase separation (LLPS). This membraneless organelle, the nucleolus, is sequestered from the nucleus by liquid droplet formation through LLPS. For the nucleolus and other subnuclear organelles, the formation and regulation of LLPS are closely associated with oncogenesis, tumor progression, and metastasis
[101][102][101,102]. Ide et al. used single-molecule tracking to show that RNA polymerase I (Pol I) and chromatin-bound upstream binding factor (UBF) undergo transcription suppression through phase separation
[39]. Active Pol I forms small clusters in the FC, restricting rDNA chromatin. The inhibition of transcription causes Pol I to disassociate from rDNA, becoming liquid-like in the nucleolar cap. A Pol I mutant linked to a craniofacial disorder competes with wild-type Pol I, transforming the FC into a cap and inhibiting transcription. The cap droplet excludes an initiation factor, ensuring effective silencing. This reveals a mechanism of rRNA transcription suppression via Pol I-mediated phase separation within the nucleolus.
Condensates induced by transcription inhibition (CITIs) in the nucleolus drastically alter the spatial organization of the genome. CITIs are formed by the splicing factor proline- and glutamine-rich (SFPQ) protein, the non-POU domain-containing octamer-binding protein (NONO), fused in sarcoma (FUS), and TATA-Box-binding protein-associated factor 15 (TAF15) in nucleoli upon the inhibition of RNA polymerase II (RNAPII). Yasuhara et al. found that the SFPQ protein and NONO undergo rapid LLPS in nucleoli upon RNAPII inhibition, resulting in the formation of CITIs
[103]. The localization of active chromatin to CITIs increases the illegitimate fusions of DNA double-strand breaks (DSBs) in active genes, promoting the formation of fusion oncogenes. It has been suggested that proper RNAPII transcription and rRNA processing are essential for preventing the LLPS of SFPQ/NONO on rRNA.