1. Introduction
Omics approaches have played a crucial role in unraveling the intricate mechanisms of diseases within biological systems. Their use has increased significantly over the past decades. At first, genomic studies were widely used in discovering genetic variations associated with certain diseases and diverse treatment outcomes. In addition to genomics, other omics technologies such as epigenomics, transcriptomics, proteomics, and metabolomics have also proven to be highly valuable in deepening
ouresearcher
s' knowledge of biochemical mechanisms
[1]. Metabolomics has a significant advantage in this scenario as it represents the final stage in the omics cascade, enabling the closest correlation to the observed disease phenotype
[2]. This omics approach employs high-throughput analytical techniques to measure the levels of low molecular weight molecules (known as metabolites) in biological samples offering a better understanding of the biochemical mechanisms and pathways associated with a diverse range of diseases
[3,4][3][4].
The variations observed among individuals in their genome, proteome, and metabolome have a profound influence on how they respond to treatment and the achieved outcomes. Thus, the concept of personalized medicine is based on the idea that individuals’ unique characteristics require tailored interventions for their specific diseases
[5]. This concept has been applied in cancer research, particularly in prostate, bladder, and breast cancers, where diverse clinical outcomes pose significant challenges in the effective treatment and management of these diseases
[6,7][6][7]. Particularly for bladder cancer (BC), personalized medicine is crucial due to the inherent heterogeneity of the disease
[8,9][8][9]. BC exhibits diverse molecular profiles and clinical behaviors among patients, making the uniform treatment approaches less effective. Moreover, lifelong BC surveillance is common due to its high recurrence, which increases costs and affects the quality of life of patients.
[6].
Several studies have used omics to find biomarkers for personalized medicine in BC
[6,10][6][10]. A proteomics study revealed a panel of cytokines to predict recurrence after intravesical immunotherapy in BC patients with 85.5% accuracy
[11]. Genomic studies, for instance, have revealed that modifications of oncogenes such as
cyclin-dependent kinase (CDK) and
fibroblast growth factor receptor (FGFR3) serve as predictive biomarkers of response to their respective inhibitors
[12,13][12][13]. Although metabolomics research in BC personalized medicine may be less prevalent compared to genomics, its potential is enormous since changes in metabolite levels can be used as biomarkers to predict drug responses
[5,14][5][14].
2. Tracking Treatment Response in BC Using Metabolomics Approaches
2.1. Metabolomics Workflow
Metabolomic approaches have been successful in detecting various types of cancer, including lung, breast, bladder, and prostate cancers
[76,77,78,79][15][16][17][18]. One of the key advantages of metabolomics is that it considers environmental factors when analyzing cellular and systemic metabolic profiles. This positioning downstream in the omics cascade allows filling the gap between the genotype and phenotype for a more comprehensive understanding of these processes. Therefore, the molecules identified in a metabolomic analysis, including amino acids, lipids, nucleotides, and organic acids, among others, hold great promise as diagnostic, prognostic, and monitoring biomarkers
[80,81][19][20]. These biomarkers can greatly assist in the personalized treatment and management of BC.
The metabolomics approach is commonly divided into untargeted and targeted analyses. The untargeted analysis consists of detecting all possible metabolites in a single analytical run, thereby providing a global metabolic profile of the samples, while the targeted analysis measures the concentrations of specific metabolites
[82,83][21][22]. The latter can be used after untargeted analysis to obtain precise quantification of the compounds detected.
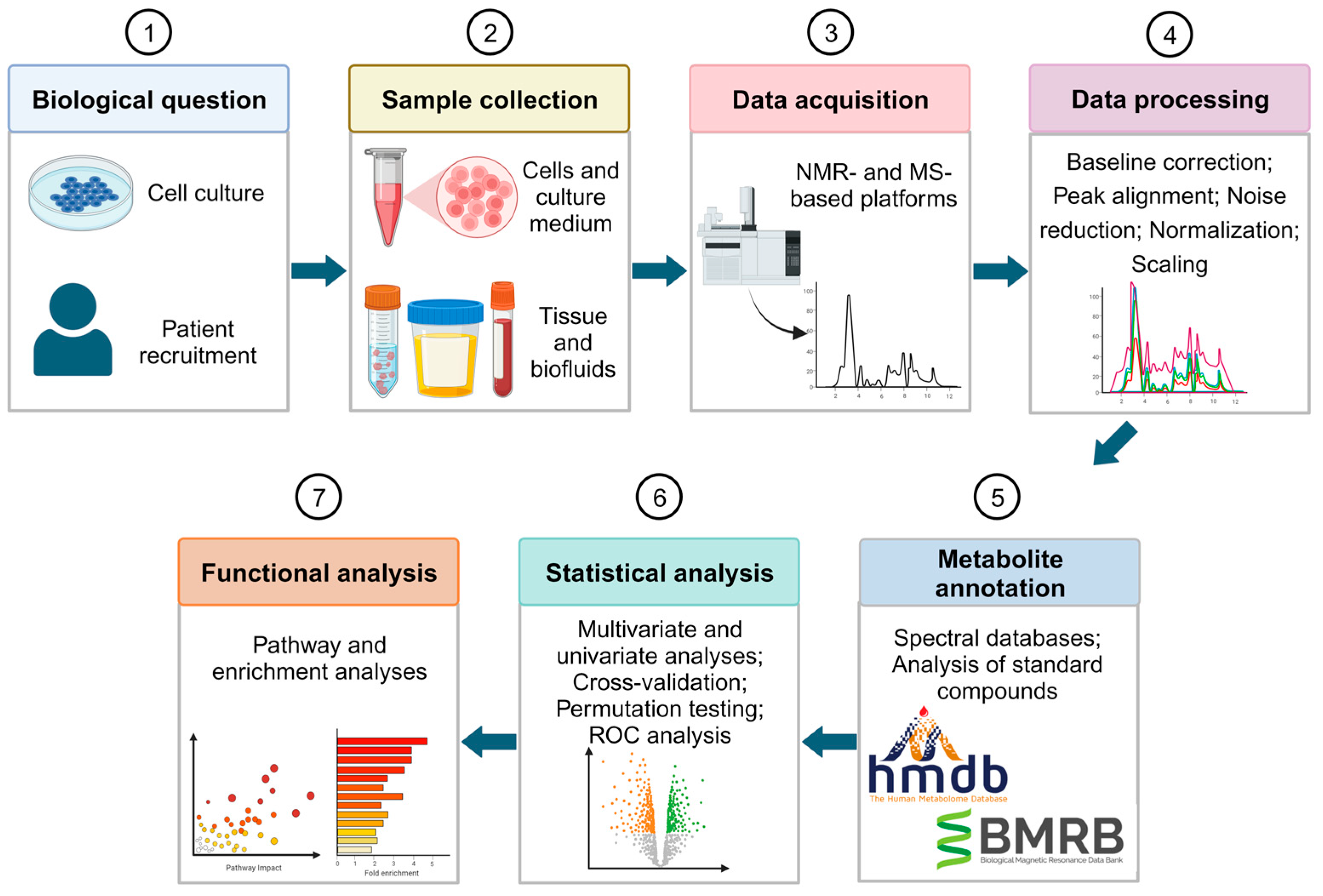
Figure 1 shows the workflow designed for an untargeted metabolomic study to find biomarkers for BC diagnosis or prognosis. First, samples are collected to analyze their metabolic profile. Samples can be collected directly from the patient either as a biofluid, such as urine and blood plasma/serum, or as tissue. Additionally, studies can be based on in vitro models. These models are useful for an initial study of the metabolic changes associated with the desirable therapy, since it is less complex, and samples have less variability
[84][23]. However, it may be difficult to extrapolate the findings of in vitro studies to in vivo systems. To achieve precise findings, the use of human tissues or biofluids is highly recommended. Moreover, biofluids have the added advantage of being non-invasive.
Figure 1. Overview of the untargeted metabolomics workflow for biomarker discovery in BC diagnosis and prognosis. Figure created using Biorender.com.
After sample collection, high-throughput analytical techniques are needed to detect and characterize the metabolites that are present in the samples. Mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy stand out as the most used analytical platforms for metabolic profiling
[82][21]. MS detects and quantifies the metabolites by measuring their mass via the mass-to-charge ratio (
m/
z). MS techniques are usually coupled to separation methods like gas chromatography (GC) and liquid chromatography (LC) to reduce sample complexity, enhance sensitivity, and facilitate metabolite annotation by providing a retention time identifier
[85][24]. MS-based metabolomics studies require the use of quality control (QC) samples to assess and monitor the reliability, reproducibility, and overall quality of the analytical process
[86][25]. In contrast, NMR relies on atomic nuclei absorption and re-emission, providing quantitative and reproducible measurements of known and unknown metabolites, requiring minimal sample preparation.
The main objective of data processing in metabolomics is to accurately identify and quantify the various features present in the data. The process involves several comparable steps for MS and NMR data including baseline correction, peak alignment, noise reduction, normalization, and scaling
[82,87,88][21][26][27]. The endpoint of data processing is a feature matrix that includes intensities or abundances of relevant signals for each sample, representing the metabolic fingerprint of each individual.
Metabolite annotation is essential in metabolomics to identify and assign chemical identities to detected metabolomic features
[89][28]. The most used method involves matching experimental data with reference data from databases or spectral libraries
[90][29]. However, additional procedures are required to obtain accurate metabolite annotation. Several researchers have focused on minimal reporting standards for levels of confidence in metabolite annotation
[91][30], which are essential for accurately evaluating the credibility of any biochemical interpretation within a metabolomics study. To validate metabolite annotations, it is recommended to use at least two independent and complementary datasets for the analysis of a chemical reference standard with suspected structural equivalence. Datasets may consist of retention time/index and mass spectrum data, accurate mass and tandem MS (MS/MS) results, as well as full
1H and/or
13C NMR data. All analyses should be carried out under identical analytical conditions in the same laboratory.
Statistical methods are employed to find metabolites that are significantly expressed, which are categorized into multivariate and univariate analyses. Multivariate analysis identifies patterns and differences between case and control groups considering a large number of variables. Some examples of multivariate methods include principal component analysis (PCA), and partial least square-discriminant analysis (PLS-DA)
[95][31]. Cross-validation and permutation testing are important in assessing the robustness of predictive models, particularly in the case of small sample sizes
[96,97][32][33]. Conversely, univariate analysis such as
t-tests, and analysis of variance (ANOVA) evaluate a single variable. Given the presence of multiple metabolites in biological samples, multiple testing corrections (e.g., Bonferroni, false discovery rate) are essential to mitigate false positives associated with univariate testing
[98][34]. In addition, biomarker discovery studies also consider receiver operator characteristic (ROC) curve analysis to evaluate the performance of biomarkers in terms of sensitivity, specificity, and accuracy
[99][35].
Functional analysis is the next step of a metabolomics study. The compounds detected in this analysis are linked to their biological pathways and functions to uncover altered biomarkers and their associations with biological and medical conditions
[98][34]. Enrichment or pathway analyses are common methods employed for this purpose
[100][36].
Ultimately, the study’s findings need validation in larger cohorts, considering training and testing sets for application in clinical practice for diagnostic, prognostic, and monitoring purposes
[101][37]. Integration with other omics data, including genomics, transcriptomics, or proteomics, further enhances the understanding of the functions and interactions of differentially expressed metabolites
[102][38].
3.2. Metabolomic Biomarkers of Treatment Response in BC
Metabolomics has the potential to predict disease progression and treatment response by monitoring metabolic changes. Thus, discovering biomarkers through metabolomics for personalized BC treatment is of great importance. This approach has the potential to decrease the impact of the illness, as it is less invasive and more cost-effective. This part presents an extensive review of studies that have used metabolomics approaches to identify potential biomarkers of treatment response in BC. The search was conducted on the PubMed database with the specified keywords: (“metabolomics” OR “metabolic profiling”) AND (transurethral resection OR chemotherapy OR immunotherapy) AND “bladder cancer”. The search included all English publications without any restrictions on the publication date and was conducted in May 2023. The search retrieved 35 papers that were screened according to the following inclusion criteria: (1) papers reporting original results; (2) papers using metabolomic approaches; (3) papers focused on current BC treatment options (transurethral resection of the bladder tumor, chemotherapy, and immunotherapy). After excluding reviews and other studies not relevant to the topic, seven studies were considered eligible, from which three were in vitro studies, three considered human biofluids (blood serum and urine), and one considered tumoral and non-tumoral tissues.
3.2.1. In Vitro Studies
MIBC patients often receive cisplatin-based chemotherapy for treatment. The primary mechanism of action of cisplatin is through DNA damage, which activates several signal transduction pathways leading to apoptosis
[103][39]. Resistance to cisplatin chemotherapy is a major limitation, but the reasons behind this resistance remain mostly unknown
[104][40]. Two in vitro studies have been conducted to analyze the metabolism of cisplatin-resistant BC cells using metabolomics
[105,106][41][42]. Another in vitro study delved into the metabolic mechanisms driving the multiple drug resistance in BC
[107][43].
Lee et al. focused on lipid metabolism to investigate the mechanisms and pathways underlying the resistance to cisplatin-based treatments
[106][42]. Changes in lipid metabolism have been associated with aggressive forms of some cancers and may be used as potential cancer biomarkers
[108][44]. For this, the authors used two transitional cell carcinoma (T24) cell lines, one cisplatin-sensitive (T24S) and one cisplatin-resistant (T24R). Through ultra-performance liquid chromatography-mass spectrometry (UPLC-MS), differentially expressed lipids were identified by comparing the T24S and T24R cells
[106][42]. The lipid profiles of T24S and T24R cells treated with and without an acetyl-CoA synthetase 2 (ACSS2) inhibitor were also examined. The purpose was to investigate the significance of this enzyme in fatty acid synthesis by analyzing T24S+ and T24R+ cells (treated with the inhibitor) in comparison to T24S− and T24R− cells (untreated). The results revealed a decrease in cisplatin-induced apoptosis and an increase in lipidic production in T24R cells, suggesting a link between the two events. Compared to T24S cells, the T24R cells displayed a high number of downregulated lipid species, including various phosphatidylethanolamines (PE) and triglycerides (TG). On the other hand, one ceramide (CE) and several phosphatidylcholines (PCs) and sphingomyelins (SMs) were found to be upregulated in T24R cells. ACSS2’s association with treatment resistance was confirmed by the authors, who found greater differences between T24S+ and T24R+ cells compared to the differences observed between T24S− and T24R− cells. ACSS2 inhibition did not significantly affect most T24R-specific lipidic metabolites, although metabolites such as CE (18:1), CE (22:6), TG (49:1), and TG (53:2) were greatly affected by the ACSS2 inhibitor. These results suggest an association between cisplatin resistance and altered lipid metabolism. The authors highlighted the potential of modulating lipid metabolism as a promising approach to combat cisplatin resistance.
Wen et al. also used T24S and T24R cell lines in their study, but instead of using an MS-based technique, they performed a real-time NMR-based metabolomics approach to detect metabolic changes in cisplatin-resistant BC cells
[105][41]. In this approach, the metabolites generated from
13C-glucose tracer were monitored by two-dimensional (2D)
1H-
13C Heteronuclear Single Quantum Coherence (HSQC) NMR in real time. A Western blot analysis was also performed to check possible alterations of fatty acid metabolism involving acetate. The T24R cells revealed higher glucose consumption, higher and faster acetate accumulation, higher levels of fatty acids and lower production and excretion of lactate. On the other hand, T24S cells showed higher and faster lactate and alanine accumulation. Thus, cisplatin resistance may be related to glucose consumption and acetate production. The results from the Western blot revealed an increase in upstream metabolic regulators in T24R cells, namely phosphorylated epidermal growth factor receptor (EGFR) and mammalian target of rapamycin (mTOR), suggesting an association of these regulators with cisplatin resistance. In addition, acetyl-CoA carboxylase and ACSS2 were more highly expressed in T24R cells. The higher levels of both enzymes may indicate that T24R cells have an enhanced fatty acid synthesis via two carbon metabolism involving acetate. Moreover, the authors confirmed that glucose-derived endogenous acetate contributes to the enhanced fatty acid de novo synthesis in T24R cells. Inhibition of the ACSS2 pathway decreased the de novo synthesis of fatty acids in T24R cells only. ACSS2 expression in bladder tumor tissues from patients receiving cisplatin-based chemotherapy was further increased, confirming the relevance of ACSS2 in cisplatin resistance.
Zhu et al. aimed to investigate the mechanism of multiple drug resistance in BC with a specific focus on exploring the involvement of both c-MYC and polyamine metabolism
[107][43]. Polyamines, as essential low molecular compounds, play a critical role in eukaryotic cell growth and function. In contrast, c-MYC is a well-known oncogene that significantly influences cell proliferation, senescence, and apoptosis
[107,109,110][43][45][46]. The authors created a pirarubicin-resistant cell line (T24/THP) with multiple drug resistance by gradually exposing the T24 cell line to increasing concentrations of pirarubicin, epirubicin, and mitomycin C. This resistant cell line was compared with the parental T24 cell line. To gain insights into the underlying mechanisms of multiple drug resistance, a combination of untargeted metabolomics analysis and gene microarray detection was employed. The researchers discovered variations in the concentrations of numerous metabolites that are associated with the metabolism of arginine and proline. These findings indicate that this pathway could potentially play a significant role in the development of resistance. Moreover, the levels of two polyamines (putrescine and spermidine), as well as c-MYC, were observed to decrease in pirarubicin-resistant cells. Although the expression of ornithine decarboxylase (ODC1) and spermidine synthetase (SRM) is low in resistant cells, their knockdown surprisingly revealed contrasting outcomes by enhancing treatment sensitivity. ODC1 and SRM may contribute to drug resistance, as their lower expression in pirarubicin-resistant cells could be influenced by other upstream genes. Furthermore, the regulation of ODC1 and SRM expression by c-MYC, as well as its impact on drug resistance in T24PR cells, remains a subject of debate due to conflicting results from different studies on the therapeutic benefits of its downregulation
[111,112][47][48].
Overall, these studies provide novel insights into the mechanisms of drug resistance in BC, offering potential targets for future therapeutic interventions. The three studies examined revealed a notable challenge in achieving reliable metabolite annotation, which is particularly evident in LC-MS investigations featuring numerous differential metabolites. This limitation poses a potential obstacle to the precise identification of critical biomarkers or therapeutic targets. Furthermore, a primary concern in these studies is the inherent challenge of extrapolating results to in vivo models, suggesting that the identified potential biomarkers (differential metabolites) may not comprehensively reflect the complex physiological processes within the human body. Despite this limitation, in vitro studies play a vital role in providing essential insights into the metabolic pathways and treatment-induced alterations specific to bladder tumor cells.
3.2.2. Human Biofluid and Tissue Studies
Investigating the metabolomic features of cancer treatment response in human subjects is crucial for enhancing
ouresearcher
s' knowledge of therapeutic outcomes and improving personalized treatment strategies. However, the number of reported studies in this field for BC is relatively limited.
OurResearchers' search yielded two studies that investigated the impact of TURBT on the metabolic profile characteristic of BC using human biofluids (blood serum and urine)
[113,114][49][50]. Additionally, another study focused on evaluating the impact of submucosal injection of gemcitabine prior to TURBT on both BC and adjacent normal tissue
[115][51], while a separate investigation aimed to identify biomarkers in blood serum for predicting the effectiveness of neoadjuvant chemotherapy in BC
[116][52].
Bansal et al. previously reported that alterations in the levels of dimethylamine (DMA), malonate, lactate, glutamine, histidine, and valine demonstrate a 95% detection rate for BC cases when compared to healthy controls
[117][53]. The same research team carried out a targeted metabolomics analysis to examine the levels of the aforementioned metabolites and their potential to differentiate between pre- and post-operative conditions in patients with BC
[113][49]. The study consisted of 130 men who were diagnosed with BC (LG
n = 33–35 and HG
n = 31) and underwent TURBT. Blood serum samples were collected before surgery (pre-operative samples) and 30, 60, and 90 days after surgery (post-operative samples). To evaluate the expression level of potential biomarkers, the authors conducted a comparison between pre- and post-operative sample signatures, which were further divided into LG and HG categories. The study also included an additional group of 52 controls. Of the selected metabolites, DMA, lactate, glutamine, histidine, and valine levels were higher in LG BC compared to controls, and these levels decreased after TURBT. Moreover, DMA, malonate, lactate, histidine, and valine also presented higher levels in HG BC compared to controls and decreased levels throughout post-operation time. These findings suggest that DMA, lactate, histidine, and valine hold promise as potential non-invasive biomarkers for inclusion in follow-up protocols for BC.
Previous research conducted by Jacyna et al. highlighted changes in the levels of various metabolites involved in amino acid, pyrimidine, purine, and energy metabolisms in the urine of BC patients in comparison to that of healthy controls
[118][54]. Later, the authors applied the same multi-platform metabolomics approach to measure levels of selected metabolites in the urine of BC patients who had undergone TURBT
[114][50]. Eight men and two women diagnosed with NMIBC participated in this study. The samples were collected before, the day after, and two weeks after the TURBT procedure. Two weeks after TURBT, the levels of several urinary metabolites were found significantly decreased in comparison with pre-TURBT, including metabolites participating in the amino acid, purine, and pyrimidine metabolisms. These molecules participate in a wide range of biological processes such as cell proliferation and tumor progression
[119,120][55][56]. In addition, several differential molecules found by Jacyna et al. are methylated metabolites possibly related with processes of DNA methylation enhanced during BC
[121][57]. In particular, the levels of 1,3-dimethyluracil, N1-methyl-2-1,3-dimethyluracil, and methylnicotinamide were significantly decreased two weeks after TURBT in comparison to pre-TURBT. These results indicate that post-surgery metabolomic-based approaches can effectively assess the eradication of the tumor’s metabolic phenotype. The study faced a significant limitation due to the small number of samples, as only 20% of the initially recruited patients attended the follow-up visit. Further research is required to precisely identify biomarkers that can be implemented in clinical settings.
The submucosal injection of gemcitabine prior to TURBT may prevent BC recurrence, but the underlying mechanism remains unknown. In this context, Yang et al. applied a MS-based metabolomic approach to analyze metabolic changes in BC and normal bladder tissues before and after treatment with gemcitabine
[115][51]. The study participants comprised a small cohort of 12 BC patients (nine men and three women), all of whom had undergone TURBT. Almost all patients were diagnosed with HG, T2 stage tumors (
n = 9), except one T1 BC female patient, and two male patients were diagnosed with Ta malignancies. First, the authors compared pre-gemcitabine normal tissues with pre-gemcitabine BC tissues indicating that changes in glutathione, purine, and thiamine metabolism pathways were significantly associated with BC malignancy. After gemcitabine treatment, the bilirubin and retinal levels found higher in BC tissues recovered to levels similar to that of pre-gemcitabine normal tissue. This result suggested that these metabolites may be the potential targets of gemcitabine for reducing BC recurrence. The submucosal injection of gemcitabine also affected the metabolic composition of the adjacent normal tissue since higher levels of histamine and lower levels of thiamine were observed in post- compared with pre-gemcitabine normal tissues. Yang et al. suggested that histamine could protect against disease relapse whereas thiamine could be related to side effects of the treatment.
Zhuang et al. employed NMR- and MS-based metabolomics methods to discover metabolic biomarkers for predicting the effectiveness of neoadjuvant chemotherapy (NAC) in MIBC patients
[116][52]. Before the first chemotherapy, blood serum was collected from 18 patients who were scheduled to receive NAC (gemcitabine and cisplatin). The patients were divided into two groups: six patients were identified as NAC-sensitive (T2
n = 4, and T3
n = 2), while the remaining 12 patients were categorized as NAC-resistant (T2
n = 5, T3
n = 6, and T4
n = 1). The analysis of serum samples was performed using both
1H NMR spectroscopy and UPLC-MS. The sera of NAC-sensitive patients displayed decreased levels of various metabolites, such as organic acids and amino acids, in contrast to NAC-resistant patients. Furthermore, there was an upregulation of glutamine and taurine in NAC-sensitive patients as opposed to NAC-resistant patients. From these metabolites, the authors emphasized the variations found in the levels of glutamine, taurine, glycine, and hypoxanthine, which were detected in both analytical methods with the same trend. The pathway analysis unveiled an enrichment of glutathione metabolism, glycine, serine and threonine metabolism as well as glyoxylic acid and dicarboxylic acid metabolism. The three putative metabolic pathways may be important for determining chemotherapy sensitivity. Although this study had limitations in terms of the number of participants, it successfully demonstrated that there are variations in metabolic phenotypes between NAC-sensitive and NAC-resistant patients. The discovery highlights the potential of metabolomics in customizing medical treatments for individuals diagnosed with MIBC.
Several limitations in the reviewed studies should be highlighted, as they hinder the robustness of treatment response biomarkers in BC. These limitations included small sample sizes, lack of robustness in metabolite annotation (mostly level 2
[91][30]), and some studies’ failure to consider both genders. The limited sample sizes may arise from challenges in patient recruitment, as certain studies involved multiple time points gathered from each participant. Moreover, the diverse range of BCa tumors, which included different stages and grades, along with diverse treatment protocols and research methodologies, pose challenges when attempting to compare different studies.