Zinc finger and BTB domain-containing 20 (ZBTB20), which was initially identified in human dendritic cells, belongs to a family of transcription factors (TFs) with an N-terminal BTB domain and one or more C-terminal DNA-binding zinc finger domains. Under physiological conditions, ZBTB20 acts as a transcriptional repressor in cellular development and differentiation, metabolism, and innate immunity. Interestingly, multiple lines of evidence from mice and human systems have revealed the importance of ZBTB20 in the pathogenesis and development of cancers. ZBTB20 is not only a hotspot of genetic variation or fusion in many types of human cancers, but also a key TF or intermediator involving in the dysregulation of cancer cells.
- ZBTB20
- transcription factor
- human
- cancer
- hematological malignancy
1. Introduction

2. Functional Domains of ZBTB Proteins
2.1. The BTB Domain
2.2. The C2H2-Type ZF Domain
2.3. Other Known Domains
2.4. The Linker Region
2.5. ZBTB Family Members
3. Physiological Roles of ZBTB20
3.1. Lymphoid Development and Differentiation
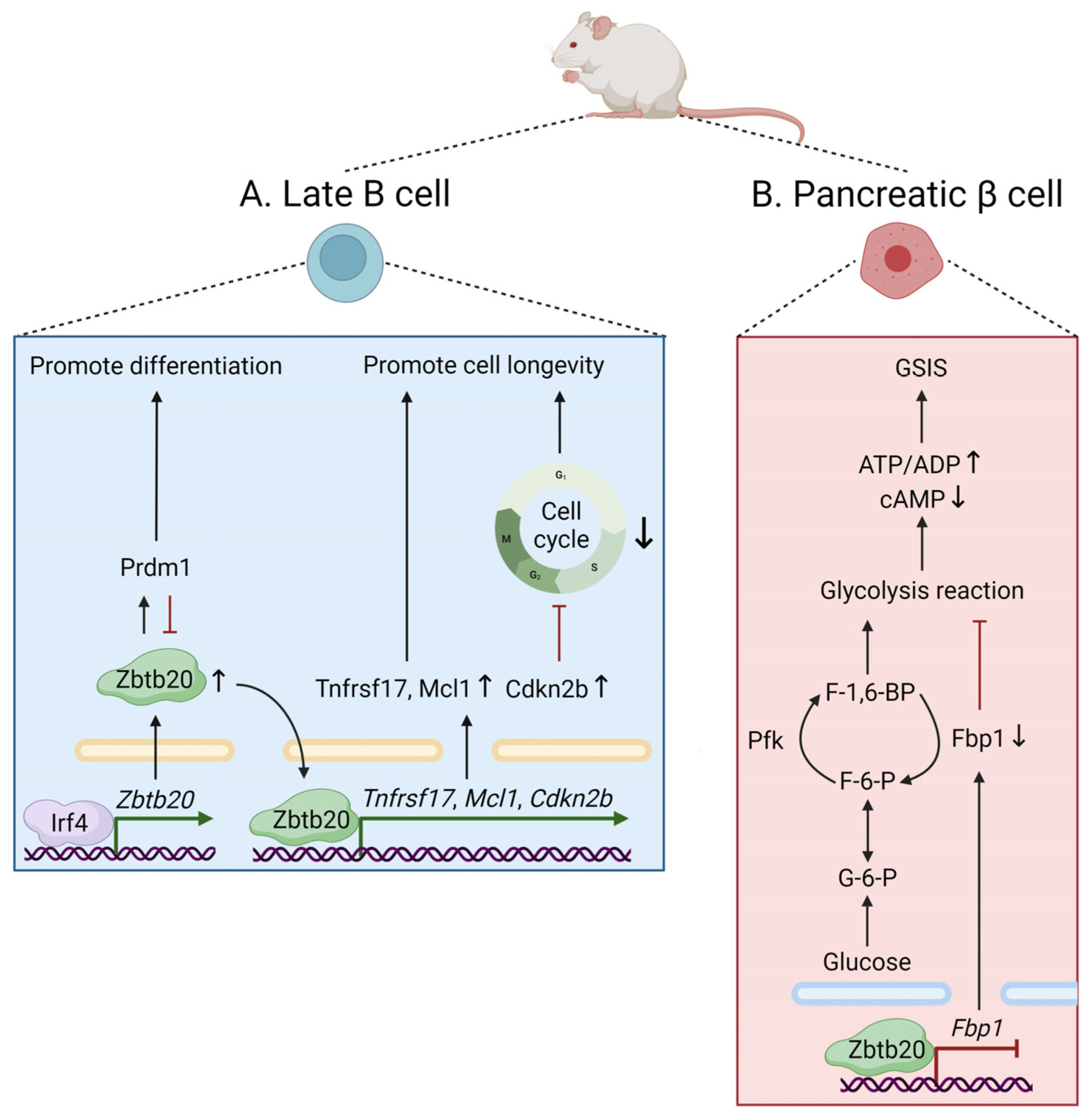
3.2. Cellular Metabolism
3.3. Neurodevelopment
3.4. Immune Response and Inflammation
4. ZBTB20 in Cancers
|
Cancer Type |
Potential Function/Mechanism |
Ref. |
||
|---|---|---|---|---|
|
HCC |
Mice |
Transcriptional repression of the Afp gene |
||
|
(1) Promotion of liver regeneration (2) Regulation of the hepatic expression of Egfr |
||||
|
One of nineteen highly significant candidate locus implicated in mouse HCC |
||||
|
Humans |
Reactivation of AFP via the miR-122-mediated regulation |
|||
|
An independent marker for poor prognosis in human HCC |
||||
|
Promotion of HCC by suppressing FOXO1 |
||||
|
Promotion of HCC by correlation with SETD7 |
||||
|
Suppression of HCV infection |
||||
|
Association with HBV integration frequency |
||||
|
GC |
Humans |
Identification of rs9841504 as a new susceptibility locus for non-cardia GC in the Chinese population |
||
|
Association of rs9841504 with severe intestinal metaplasia/atypical hyperplasia in the Chinese population |
||||
|
Identification of rs758277701 in the MSI subtype of GC in the Korean population |
||||
|
Identification of rs9288999 as a protective factor for reducing GC risk in the Chinese Han population |
||||
|
Promotion of GC via the NFKBIA/NF-κB signaling pathway |
||||
|
CNS cancer |
Humans |
Glioma |
A mutation hotspot |
|
|
Neuronal and mixed neuronal-glial tumors |
Implication of miRNAs that target ZBTB20 in the classification of pediatric cases |
|||
|
Low-grade glioma |
Identification of ZBTB20-AS4 as a critical IncRNA for predicting the prognosis |
|||
|
Shh-MB |
Identification of the fusion transcripts of ZBTB20 as recurrent fusions |
|||
|
Glioblastoma |
Promotion of glioblastoma through the miR-758-5p/ZBTB20 axis or by ZBTB20 |
|||
|
Blood cancer |
Humans |
B-CLL |
The top differentially expressed gene in terms of VH mutation status |
|
|
AML |
Promotion of cell growth and migration via the LINC00641/miR-378a/ZBTB20 axis |
|||
|
Promotion of malignant phenotypes via the circ-SFMBT2/miR-582-3p/ZBTB20 axis |
||||
|
Promotion of leukemia development via the circ-0001602/miR-192-5p/ZBTB20 axis |
||||
|
MCL |
(1) A novel downstream target repressed by BACH1 (2) Involvement in the BACH1-mediated regulation of tumor immune microenvironment |
|||
|
Others |
Humans |
BC |
Downregulation in ERα+ BC biopsies upon treatment of aromatase inhibitors |
|
|
Upregulation in ERα+ BC cell lines upon anacardic acid treatment |
||||
|
Promotion of cell migration and invasion via the SNHG8/miR-634/ZBTB20 axis |
||||
|
Promotion of cell proliferation, migration, and invasion via the circ-0104345/miR-876-3p/ZBTB20 axis |
||||
|
Colorectal cancer |
Identification of rs10511330 and rs16822593 as two of the top 10 SNPs in patients |
|||
|
Cervical cancer |
One of ten potential driver genes |
|||
|
NSCLC |
(1) Upregulation in NSCLC tissues (2) Promotion of cell proliferation by repressing FOXO1 |
|||
|
Ovarian cancer |
Increase in cells that migrate in omentum tissue pretreated with extracellular vesicles isolated from ascitic supernatant of high-grade patients |
|||
Note: AML, acute myeloid leukemia; AFP/Afp, alpha-fetoprotein; BACH1, BTB and CNC homology 1; BC, breast cancer; B-CLL, B-cell chronic lymphoblastic leukemia; circ-SFMBT2, circular RNA Scm-like with four mbt domains 2; Egfr, epithelial growth factor receptor; ERα+, estrogen receptor α positive; FOXO1, forkhead box O1; GC, gastric cancer; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; IncRNA, long non-coding RNA; miRNA, microRNA; MCL, mantle cell lymphoma; MSI, microsatellite instability; NSCLC, non-small cell lung cancer; Ref., references; SETD7, SET domain-containing 7; Shh-MB, Sonic Hedgehog medulloblastoma; SNHG8, small nucleolus RNA host gene 8; SNPs, single nucleotide polymorphisms.
References
- Maeda, T. Regulation of hematopoietic development by ZBTB transcription factors. Int. J. Hematol. 2016, 104, 310–323.
- Zhang, W.; Mi, J.; Li, N.; Sui, L.; Wan, T.; Zhang, J.; Chen, T.; Cao, X. Identification and characterization of DPZF, a novel human BTB/POZ zinc finger protein sharing homology to BCL-6. Biochem. Biophys. Res. Commun. 2001, 282, 1067–1073.
- Sutherland, A.P.; Zhang, H.; Zhang, Y.; Michaud, M.; Xie, Z.; Patti, M.E.; Grusby, M.J.; Zhang, W.J. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol. Cell. Biol. 2009, 29, 2804–2815.
- Rosenthal, E.H.; Tonchev, A.B.; Stoykova, A.; Chowdhury, K. Regulation of archicortical arealization by the transcription factor Zbtb20. Hippocampus 2012, 22, 2144–2156.
- Zhang, Y.; Xie, Z.; Zhou, L.; Li, L.; Zhang, H.; Zhou, G.; Ma, X.; Herrera, P.L.; Liu, Z.; Grusby, M.J.; et al. The zinc finger protein ZBTB20 regulates transcription of fructose-1,6-bisphosphatase 1 and beta cell function in mice. Gastroenterology 2012, 142, 1571–1580.e6.
- Liu, X.; Zhang, P.; Bao, Y.; Han, Y.; Wang, Y.; Zhang, Q.; Zhan, Z.; Meng, J.; Li, Y.; Li, N.; et al. Zinc finger protein ZBTB20 promotes Toll-like receptor-triggered innate immune responses by repressing IκBα gene transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 11097–11102.
- Chevrier, S.; Emslie, D.; Shi, W.; Kratina, T.; Wellard, C.; Karnowski, A.; Erikci, E.; Smyth, G.K.; Chowdhury, K.; Tarlinton, D.; et al. The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. J. Exp. Med. 2014, 211, 827–840.
- Wang, Y.; Bhattacharya, D. Adjuvant-specific regulation of long-term antibody responses by ZBTB20. J. Exp. Med. 2014, 211, 841–856.
- Sun, Y.; Preiss, N.K.; Valenteros, K.B.; Kamal, Y.; Usherwood, Y.K.; Frost, H.R.; Usherwood, E.J. Zbtb20 Restrains CD8 T Cell Immunometabolism and Restricts Memory Differentiation and Antitumor Immunity. J. Immunol. 2020, 205, 2649–2666.
- Consortium, T.U. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022, 51, D523–D531.
- Bardwell, V.J.; Treisman, R. The POZ domain: A conserved protein-protein interaction motif. Genes Dev. 1994, 8, 1664–1677.
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.; Nandra, S.K.; Prive, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82.
- Perez-Torrado, R.; Yamada, D.; Defossez, P.A. Born to bind: The BTB protein-protein interaction domain. Bioessays 2006, 28, 1194–1202.
- Bonchuk, A.; Denisov, S.; Georgiev, P.; Maksimenko, O. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J. Mol. Biol. 2011, 412, 423–436.
- Hong, S.H.; David, G.; Wong, C.W.; Dejean, A.; Privalsky, M.L. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 1997, 94, 9028–9033.
- Ahmad, K.F.; Melnick, A.; Lax, S.; Bouchard, D.; Liu, J.; Kiang, C.L.; Mayer, S.; Takahashi, S.; Licht, J.D.; Prive, G.G. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell 2003, 12, 1551–1564.
- Seyfert, V.L.; Allman, D.; He, Y.; Staudt, L.M. Transcriptional repression by the proto-oncogene BCL-6. Oncogene 1996, 12, 2331–2342.
- Chang, C.C.; Ye, B.H.; Chaganti, R.S.; Dalla-Favera, R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA 1996, 93, 6947–6952.
- Melnick, A.; Ahmad, K.F.; Arai, S.; Polinger, A.; Ball, H.; Borden, K.L.; Carlile, G.W.; Prive, G.G.; Licht, J.D. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 2000, 20, 6550–6567.
- Kang, M.I.; Kobayashi, A.; Wakabayashi, N.; Kim, S.G.; Yamamoto, M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA 2004, 101, 2046–2051.
- Pintard, L.; Willis, J.H.; Willems, A.; Johnson, J.L.; Srayko, M.; Kurz, T.; Glaser, S.; Mains, P.E.; Tyers, M.; Bowerman, B.; et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 2003, 425, 311–316.
- Xu, L.; Wei, Y.; Reboul, J.; Vaglio, P.; Shin, T.H.; Vidal, M.; Elledge, S.J.; Harper, J.W. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 2003, 425, 316–321.
- Geyer, R.; Wee, S.; Anderson, S.; Yates, J.; Wolf, D.A. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 2003, 12, 783–790.
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139.
- Chaharbakhshi, E.; Jemc, J.C. Broad-complex, tramtrack, and bric-a-brac (BTB) proteins: Critical regulators of development. Genesis 2016, 54, 505–518.
- Cerchietti, L.C.; Ghetu, A.F.; Zhu, X.; Da Silva, G.F.; Zhong, S.; Matthews, M.; Bunting, K.L.; Polo, J.M.; Fares, C.; Arrowsmith, C.H.; et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 2010, 17, 400–411.
- Sakamoto, K.; Sogabe, S.; Kamada, Y.; Sakai, N.; Asano, K.; Yoshimatsu, M.; Ida, K.; Imaeda, Y.; Sakamoto, J.I. Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship. Biochem. Biophys. Res. Commun. 2017, 482, 310–316.
- McCoull, W.; Cheung, T.; Anderson, E.; Barton, P.; Burgess, J.; Byth, K.; Cao, Q.; Castaldi, M.P.; Chen, H.; Chiarparin, E.; et al. Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC To Provide Insight into Small Molecule Targeting of BCL6. ACS Chem. Biol. 2018, 13, 3131–3141.
- Slabicki, M.; Yoon, H.; Koeppel, J.; Nitsch, L.; Roy Burman, S.S.; Di Genua, C.; Donovan, K.A.; Sperling, A.S.; Hunkeler, M.; Tsai, J.M.; et al. Small-molecule-induced polymerization triggers degradation of BCL6. Nature 2020, 588, 164–168.
- Ai, Y.; Hwang, L.; MacKerell, A.D., Jr.; Melnick, A.; Xue, F. Progress toward B-Cell Lymphoma 6 BTB Domain Inhibitors for the Treatment of Diffuse Large B-Cell Lymphoma and Beyond. J. Med. Chem. 2021, 64, 4333–4358.
- Zacharchenko, T.; Kalverda, A.P.; Wright, S.C. Structural basis of Apt48 inhibition of the BCL6 BTB domain. Structure 2022, 30, 396–407.e3.
- Fedotova, A.A.; Bonchuk, A.N.; Mogila, V.A.; Georgiev, P.G. C2H2 Zinc Finger Proteins: The Largest but Poorly Explored Family of Higher Eukaryotic Transcription Factors. Acta Nat. 2017, 9, 47–58.
- Schmitges, F.W.; Radovani, E.; Najafabadi, H.S.; Barazandeh, M.; Campitelli, L.F.; Yin, Y.; Jolma, A.; Zhong, G.; Guo, H.; Kanagalingam, T.; et al. Multiparameter functional diversity of human C2H2 zinc finger proteins. Genome Res. 2016, 26, 1742–1752.
- Wolfe, S.A.; Nekludova, L.; Pabo, C.O. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 183–212.
- Tsuzuki, S.; Enver, T. Interactions of GATA-2 with the promyelocytic leukemia zinc finger (PLZF) protein, its homologue FAZF, and the t(11;17)-generated PLZF-retinoic acid receptor α oncoprotein. Blood 2002, 99, 3404–3410.
- Guidez, F.; Howell, L.; Isalan, M.; Cebrat, M.; Alani, R.M.; Ivins, S.; Hormaeche, I.; McConnell, M.J.; Pierce, S.; Cole, P.A.; et al. Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 2005, 25, 5552–5566.
- Donaldson, N.S.; Daniel, Y.; Kelly, K.F.; Graham, M.; Daniel, J.M. Nuclear trafficking of the POZ-ZF protein Znf131. Biochim. Biophys. Acta 2007, 1773, 546–555.
- Reeves, R.; Nissen, M.S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990, 265, 8573–8582.
- Huth, J.R.; Bewley, C.A.; Nissen, M.S.; Evans, J.N.; Reeves, R.; Gronenborn, A.M.; Clore, G.M. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat. Struct. Biol. 1997, 4, 657–665.
- Aktar, S.; Sasaki, H.; Unoki, M. Identification of ZBTB24 protein domains and motifs for heterochromatin localization and transcriptional activation. Genes Cells 2019, 24, 746–755.
- Fedele, M.; Benvenuto, G.; Pero, R.; Majello, B.; Battista, S.; Lembo, F.; Vollono, E.; Day, P.M.; Santoro, M.; Lania, L.; et al. A novel member of the BTB/POZ family, PATZ, associates with the RNF4 RING finger protein and acts as a transcriptional repressor. J. Biol. Chem. 2000, 275, 7894–7901.
- Dent, A.L.; Shaffer, A.L.; Yu, X.; Allman, D.; Staudt, L.M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 1997, 276, 589–592.
- Ye, B.H.; Cattoretti, G.; Shen, Q.; Zhang, J.; Hawe, N.; de Waard, R.; Leung, C.; Nouri-Shirazi, M.; Orazi, A.; Chaganti, R.S.; et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997, 16, 161–170.
- Fukuda, T.; Yoshida, T.; Okada, S.; Hatano, M.; Miki, T.; Ishibashi, K.; Okabe, S.; Koseki, H.; Hirosawa, S.; Taniguchi, M.; et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 1997, 186, 439–448.
- Johnston, R.J.; Poholek, A.C.; DiToro, D.; Yusuf, I.; Eto, D.; Barnett, B.; Dent, A.L.; Craft, J.; Crotty, S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009, 325, 1006–1010.
- Nurieva, R.I.; Chung, Y.; Martinez, G.J.; Yang, X.O.; Tanaka, S.; Matskevitch, T.D.; Wang, Y.H.; Dong, C. Bcl6 mediates the development of T follicular helper cells. Science 2009, 325, 1001–1005.
- Kovalovsky, D.; Uche, O.U.; Eladad, S.; Hobbs, R.M.; Yi, W.; Alonzo, E.; Chua, K.; Eidson, M.; Kim, H.J.; Im, J.S.; et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 2008, 9, 1055–1064.
- Savage, A.K.; Constantinides, M.G.; Han, J.; Picard, D.; Martin, E.; Li, B.; Lantz, O.; Bendelac, A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 2008, 29, 391–403.
- Jash, A.; Wang, Y.; Weisel, F.J.; Scharer, C.D.; Boss, J.M.; Shlomchik, M.J.; Bhattacharya, D. ZBTB32 Restricts the Duration of Memory B Cell Recall Responses. J. Immunol. 2016, 197, 1159–1168.
- Beaulieu, A.M.; Zawislak, C.L.; Nakayama, T.; Sun, J.C. The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat. Immunol. 2014, 15, 546–553.
- Licht, J.D.; Chomienne, C.; Goy, A.; Chen, A.; Scott, A.A.; Head, D.R.; Michaux, J.L.; Wu, Y.; DeBlasio, A.; Miller, W.H., Jr.; et al. Clinical and molecular characterization of a rare syndrome of acute promyelocytic leukemia associated with translocation (11;17). Blood 1995, 85, 1083–1094.
- Cattoretti, G.; Pasqualucci, L.; Ballon, G.; Tam, W.; Nandula, S.V.; Shen, Q.; Mo, T.; Murty, V.V.; Dalla-Favera, R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 2005, 7, 445–455.
- Duy, C.; Hurtz, C.; Shojaee, S.; Cerchietti, L.; Geng, H.; Swaminathan, S.; Klemm, L.; Kweon, S.M.; Nahar, R.; Braig, M.; et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature 2011, 473, 384–388.
- McLachlan, T.; Matthews, W.C.; Jackson, E.R.; Staudt, D.E.; Douglas, A.M.; Findlay, I.J.; Persson, M.L.; Duchatel, R.J.; Mannan, A.; Germon, Z.P.; et al. B-cell Lymphoma 6 (BCL6): From Master Regulator of Humoral Immunity to Oncogenic Driver in Pediatric Cancers. Mol. Cancer Res. 2022, 20, 1711–1723.
- Barna, M.; Pandolfi, P.P.; Niswander, L. Gli3 and Plzf cooperate in proximal limb patterning at early stages of limb development. Nature 2005, 436, 277–281.
- Fischer, S.; Kohlhase, J.; Bohm, D.; Schweiger, B.; Hoffmann, D.; Heitmann, M.; Horsthemke, B.; Wieczorek, D. Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J. Med. Genet. 2008, 45, 731–737.
- Cui, Y.; Zhou, M.; He, Q.; He, Z. Zbtb40 Deficiency Leads to Morphological and Phenotypic Abnormalities of Spermatocytes and Spermatozoa and Causes Male Infertility. Cells 2023, 12, 1264.
- Buaas, F.W.; Kirsh, A.L.; Sharma, M.; McLean, D.J.; Morris, J.L.; Griswold, M.D.; de Rooij, D.G.; Braun, R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004, 36, 647–652.
- Hobbs, R.M.; Seandel, M.; Falciatori, I.; Rafii, S.; Pandolfi, P.P. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell 2010, 142, 468–479.
- Hu, S.; Fambrough, D.; Atashi, J.R.; Goodman, C.S.; Crews, S.T. The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev. 1995, 9, 2936–2948.
- Zhu, S.; Lin, S.; Kao, C.F.; Awasaki, T.; Chiang, A.S.; Lee, T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell 2006, 127, 409–422.
- Xie, Z.; Ma, X.; Ji, W.; Zhou, G.; Lu, Y.; Xiang, Z.; Wang, Y.X.; Zhang, L.; Hu, Y.; Ding, Y.Q.; et al. Zbtb20 is essential for the specification of CA1 field identity in the developing hippocampus. Proc. Natl. Acad. Sci. USA 2010, 107, 6510–6515.
- Mitchelmore, C.; Kjaerulff, K.M.; Pedersen, H.C.; Nielsen, J.V.; Rasmussen, T.E.; Fisker, M.F.; Finsen, B.; Pedersen, K.M.; Jensen, N.A. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J. Biol. Chem. 2002, 277, 7598–7609.
- Yang, S.Y.; Long, J.; Huang, M.X.; Luo, P.Y.; Bian, Z.H.; Xu, Y.F.; Wang, C.B.; Yang, S.H.; Li, L.; Selmi, C.; et al. Characterization of Organ-Specific Regulatory B Cells Using Single-Cell RNA Sequencing. Front. Immunol. 2021, 12, 711980.
- Krzyzanowska, A.K.; Haynes Ii, R.A.H.; Kovalovsky, D.; Lin, H.C.; Osorio, L.; Edelblum, K.L.; Corcoran, L.M.; Rabson, A.B.; Denzin, L.K.; Sant’Angelo, D.B. Zbtb20 identifies and controls a thymus-derived population of regulatory T cells that play a role in intestinal homeostasis. Sci. Immunol. 2022, 7, eabf3717.
- Li, H.; Liu, G.; Wan, X.; Zhou, L.; Qin, Z.B.; Ma, X.H.; Su, K.; Liu, Y.J.; Yuan, J.; Wei, C.C.; et al. The zinc finger and BTB domain containing protein ZBTB20 regulates plasma triglyceride metabolism by repressing lipoprotein lipase gene transcription in hepatocytes. Hepatology 2022, 75, 1169–1180.
- Cordeddu, V.; Redeker, B.; Stellacci, E.; Jongejan, A.; Fragale, A.; Bradley, T.E.; Anselmi, M.; Ciolfi, A.; Cecchetti, S.; Muto, V.; et al. Mutations in ZBTB20 cause Primrose syndrome. Nat. Genet. 2014, 46, 815–817.
- Melis, D.; Carvalho, D.; Barbaro-Dieber, T.; Espay, A.J.; Gambello, M.J.; Gener, B.; Gerkes, E.; Hitzert, M.M.; Hove, H.B.; Jansen, S.; et al. Primrose syndrome: Characterization of the phenotype in 42 patients. Clin. Genet. 2020, 97, 890–901.
- Nielsen, J.V.; Nielsen, F.H.; Ismail, R.; Noraberg, J.; Jensen, N.A. Hippocampus-like corticoneurogenesis induced by two isoforms of the BTB-zinc finger gene Zbtb20 in mice. Development 2007, 134, 1133–1140.
- Tonchev, A.B.; Tuoc, T.C.; Rosenthal, E.H.; Studer, M.; Stoykova, A. Zbtb20 modulates the sequential generation of neuronal layers in developing cortex. Mol. Brain 2016, 9, 65.
- Ren, A.; Zhang, H.; Xie, Z.; Ma, X.; Ji, W.; He, D.Z.; Yuan, W.; Ding, Y.Q.; Zhang, X.H.; Zhang, W.J. Regulation of hippocampus-dependent memory by the zinc finger protein Zbtb20 in mature CA1 neurons. J. Physiol. 2012, 590, 4917–4932.
- Cao, D.; Ma, X.; Cai, J.; Luan, J.; Liu, A.J.; Yang, R.; Cao, Y.; Zhu, X.; Zhang, H.; Chen, Y.X.; et al. ZBTB20 is required for anterior pituitary development and lactotrope specification. Nat. Commun. 2016, 7, 11121.
- Wang, L.; Pittman, K.J.; Barker, J.R.; Salinas, R.E.; Stanaway, I.B.; Williams, G.D.; Carroll, R.J.; Balmat, T.; Ingham, A.; Gopalakrishnan, A.M.; et al. An Atlas of Genetic Variation Linking Pathogen-Induced Cellular Traits to Human Disease. Cell Host Microbe 2018, 24, 308–323.e306.
- Lu, L.; Shi, M.; Qiu, J.; Shi, Z.; Wang, C.; Fu, Y.; Lin, C.; Zhang, L.; Tao, J.; Liu, C.; et al. ZBTB20 regulates cardiac allograft rejection through NFsmall ka, CyrillicB-mediated inflammation in mouse heart transplantation. Transpl. Immunol. 2022, 74, 101676.
- Liu, B.; Liu, N.; Zhu, X.; Yang, L.; Ye, B.; Li, H.; Zhu, P.; Lu, T.; Tian, Y.; Fan, Z. Circular RNA circZbtb20 maintains ILC3 homeostasis and function via Alkbh5-dependent m(6)A demethylation of Nr4a1 mRNA. Cell Mol. Immunol. 2021, 18, 1412–1424.
- Xie, Z.; Zhang, H.; Tsai, W.; Zhang, Y.; Du, Y.; Zhong, J.; Szpirer, C.; Zhu, M.; Cao, X.; Barton, M.C.; et al. Zinc finger protein ZBTB20 is a key repressor of α-fetoprotein gene transcription in liver. Proc. Natl. Acad. Sci. USA 2008, 105, 10859–10864.
- Zhang, H.; Cao, D.; Zhou, L.; Zhang, Y.; Guo, X.; Li, H.; Chen, Y.; Spear, B.T.; Wu, J.W.; Xie, Z.; et al. ZBTB20 is a sequence-specific transcriptional repressor of α-fetoprotein gene. Sci. Rep. 2015, 5, 11979.
- Zhang, H.; Shi, J.H.; Jiang, H.; Wang, K.; Lu, J.Y.; Jiang, X.; Ma, X.; Chen, Y.X.; Ren, A.J.; Zheng, J.; et al. ZBTB20 regulates EGFR expression and hepatocyte proliferation in mouse liver regeneration. Cell Death Dis. 2018, 9, 462.
- Weng, M.Z.; Zhuang, P.Y.; Hei, Z.Y.; Lin, P.Y.; Chen, Z.S.; Liu, Y.B.; Quan, Z.W.; Tang, Z.H. ZBTB20 is involved in liver regeneration after partial hepatectomy in mouse. Hepatobiliary Pancreat. Dis. Int. 2014, 13, 48–54.
- Keng, V.W.; Villanueva, A.; Chiang, D.Y.; Dupuy, A.J.; Ryan, B.J.; Matise, I.; Silverstein, K.A.; Sarver, A.; Starr, T.K.; Akagi, K.; et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 2009, 27, 264–274.
- Kojima, K.; Takata, A.; Vadnais, C.; Otsuka, M.; Yoshikawa, T.; Akanuma, M.; Kondo, Y.; Kang, Y.J.; Kishikawa, T.; Kato, N.; et al. MicroRNA122 is a key regulator of α-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat. Commun. 2011, 2, 338.
- Wang, Q.; Tan, Y.X.; Ren, Y.B.; Dong, L.W.; Xie, Z.F.; Tang, L.; Cao, D.; Zhang, W.P.; Hu, H.P.; Wang, H.Y. Zinc finger protein ZBTB20 expression is increased in hepatocellular carcinoma and associated with poor prognosis. BMC Cancer 2011, 11, 271.
- Kan, H.; Huang, Y.; Li, X.; Liu, D.; Chen, J.; Shu, M. Zinc finger protein ZBTB20 is an independent prognostic marker and promotes tumor growth of human hepatocellular carcinoma by repressing FoxO1. Oncotarget 2016, 7, 14336–14349.
- Chen, Y.; Yang, S.; Hu, J.; Yu, C.; He, M.; Cai, Z. Increased Expression of SETD7 Promotes Cell Proliferation by Regulating Cell Cycle and Indicates Poor Prognosis in Hepatocellular Carcinoma. PLoS ONE 2016, 11, e0154939.
- He, Z.; Zhu, J.; Mo, J.; Zhao, H.; Chen, Q. HBV DNA integrates into upregulated ZBTB20 in patients with hepatocellular carcinoma. Mol. Med. Rep. 2020, 22, 380–386.
- Shi, Y.; Hu, Z.; Wu, C.; Dai, J.; Li, H.; Dong, J.; Wang, M.; Miao, X.; Zhou, Y.; Lu, F.; et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat. Genet. 2011, 43, 1215–1218.
- Cai, M.; Dai, S.; Chen, W.; Xia, C.; Lu, L.; Dai, S.; Qi, J.; Wang, M.; Wang, M.; Zhou, L.; et al. Environmental factors, seven GWAS-identified susceptibility loci, and risk of gastric cancer and its precursors in a Chinese population. Cancer Med. 2017, 6, 708–720.
- Kim, Y.; Cho, M.Y.; Kim, J.; Kim, S.N.; Oh, S.C.; Lee, K.A. Profiling cancer-associated genetic alterations and molecular classification of cancer in Korean gastric cancer patients. Oncotarget 2017, 8, 69888–69905.
- Bai, F.; Xiao, K. Prediction of gastric cancer risk: Association between ZBTB20 genetic variance and gastric cancer risk in Chinese Han population. Biosci. Rep. 2020, 40, BSR20202102.
- Zhang, Y.; Zhou, X.; Zhang, M.; Cheng, L.; Zhang, Y.; Wang, X. ZBTB20 promotes cell migration and invasion of gastric cancer by inhibiting IkappaBα to induce NF-kappaB activation. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3862–3872.
- Zhang, C.; Cheng, W.; Ren, X.; Wang, Z.; Liu, X.; Li, G.; Han, S.; Jiang, T.; Wu, A. Tumor Purity as an Underlying Key Factor in Glioma. Clin. Cancer Res. 2017, 23, 6279–6291.
- Huang, Y.; Gao, X.; Yang, E.; Yue, K.; Cao, Y.; Zhao, B.; Zhang, H.; Dai, S.; Zhang, L.; Luo, P.; et al. Top-down stepwise refinement identifies coding and noncoding RNA-associated epigenetic regulatory maps in malignant glioma. J. Cell. Mol. Med. 2022, 26, 2230–2250.
- Zakrzewska, M.; Gruszka, R.; Stawiski, K.; Fendler, W.; Kordacka, J.; Grajkowska, W.; Daszkiewicz, P.; Liberski, P.P.; Zakrzewski, K. Expression-based decision tree model reveals distinct microRNA expression pattern in pediatric neuronal and mixed neuronal-glial tumors. BMC Cancer 2019, 19, 544.
- Xiang, Z.; Chen, X.; Lv, Q.; Peng, X. A Novel Inflammatory lncRNAs Prognostic Signature for Predicting the Prognosis of Low-Grade Glioma Patients. Front. Genet. 2021, 12, 697819.
- Zhang, J.; Wang, N.; Wu, J.; Gao, X.; Zhao, H.; Liu, Z.; Yan, X.; Dong, J.; Wang, F.; Ba, Y.; et al. 5-Methylcytosine Related LncRNAs Reveal Immune Characteristics, Predict Prognosis and Oncology Treatment Outcome in Lower-Grade Gliomas. Front. Immunol. 2022, 13, 844778.
- Skowron, P.; Farooq, H.; Cavalli, F.M.G.; Morrissy, A.S.; Ly, M.; Hendrikse, L.D.; Wang, E.Y.; Djambazian, H.; Zhu, H.; Mungall, K.L.; et al. The transcriptional landscape of Shh medulloblastoma. Nat. Commun. 2021, 12, 1749.
- Liu, J.; Jiang, J.; Hui, X.; Wang, W.; Fang, D.; Ding, L. Mir-758-5p Suppresses Glioblastoma Proliferation, Migration and Invasion by Targeting ZBTB20. Cell. Physiol. Biochem. 2018, 48, 2074–2083.
- Haslinger, C.; Schweifer, N.; Stilgenbauer, S.; Dohner, H.; Lichter, P.; Kraut, N.; Stratowa, C.; Abseher, R. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J. Clin. Oncol. 2004, 22, 3937–3949.
- Wang, J.; Liu, Z.H.; Yu, L.J. Long non-coding RNA LINC00641 promotes cell growth and migration through modulating miR-378a/ZBTB20 axis in acute myeloid leukemia. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7498–7509.
- Chang, W.; Shang, Z.; Ming, X.; Wu, J.; Xiao, Y. Circ-SFMBT2 facilitates the malignant growth of acute myeloid leukemia cells by modulating miR-582-3p/ZBTB20 pathway. Histol. Histopathol. 2022, 37, 137–149.
- Wu, W.; Deng, J.; Chen, C.; Ma, X.; Yu, L.; Chen, L. Circ_0001602 aggravates the progression of acute myeloid leukemia by regulating the miR-192-5p/ZBTB20 axis. Hematology 2023, 28, 2240133.
- Li, G.; Feng, M.; Zhang, Z.; Liu, J.; Zhang, H. BACH1 loss exerts antitumor effects on mantle cell lymphoma cells via inducing a tumor-intrinsic innate immune response and cell cycle arrest. Mol. Cancer Res. 2023, 21, 1274–1287.
- Mackay, A.; Urruticoechea, A.; Dixon, J.M.; Dexter, T.; Fenwick, K.; Ashworth, A.; Drury, S.; Larionov, A.; Young, O.; White, S.; et al. Molecular response to aromatase inhibitor treatment in primary breast cancer. Breast Cancer Res. 2007, 9, R37.
- Schultz, D.J.; Krishna, A.; Vittitow, S.L.; Alizadeh-Rad, N.; Muluhngwi, P.; Rouchka, E.C.; Klinge, C.M. Transcriptomic response of breast cancer cells to anacardic acid. Sci. Rep. 2018, 8, 8063.
- Fan, D.; Qiu, B.; Yang, X.J.; Tang, H.L.; Peng, S.J.; Yang, P.; Dong, Y.M.; Yang, L.; Bao, G.Q.; Zhao, H.D. LncRNA SNHG8 promotes cell migration and invasion in breast cancer cell through miR-634/ZBTB20 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11639–11649.
- Wu, H.; Wang, A.; Wang, L.; Shi, F.; Lin, F.; Cui, H. A Novel circ_0104345/miR-876-3p/ZBTB20 Axis Regulates the Proliferation, Migration, Invasion, and Apoptosis of Breast Cancer Cells. Biochem. Genet. 2023, 61, 2548–2565.
- Penney, M.E.; Parfrey, P.S.; Savas, S.; Yilmaz, Y.E. Associations of single nucleotide polymorphisms with mucinous colorectal cancer: Genome-wide common variant and gene-based rare variant analyses. Biomark. Res. 2018, 6, 17.
- Xu, Y.; Luo, H.; Hu, Q.; Zhu, H. Identification of Potential Driver Genes Based on Multi-Genomic Data in Cervical Cancer. Front. Genet. 2021, 12, 598304.
- Zhao, J.G.; Ren, K.M.; Tang, J. Zinc finger protein ZBTB20 promotes cell proliferation in non-small cell lung cancer through repression of FoxO1. FEBS Lett. 2014, 588, 4536–4542.
- Mitra, A.; Yoshida-Court, K.; Solley, T.N.; Mikkelson, M.; Yeung, C.L.A.; Nick, A.; Lu, K.; Klopp, A.H. Extracellular vesicles derived from ascitic fluid enhance growth and migration of ovarian cancer cells. Sci. Rep. 2021, 11, 9149.
