Clostridioides difficile is an anaerobic, spore-forming bacterium associated with intestinal infection, manifesting a broad spectrum of gastrointestinal symptoms, ranging from mild diarrhea to severe colitis. A primary risk factor for the development of C. difficile infection (CDI) is antibiotic exposure. Elderly and immunocompromised individuals are particularly vulnerable to CDI. A pivotal aspect for comprehending the complexities of this infection relies on the utilization of experimental models that mimic human CDI transmission, pathogenesis, and progression. These models offer invaluable insights into host–pathogen interactions and disease dynamics, and serve as essential tools for testing potential therapeutic approaches.
- Clostridioides difficile
- gut
- colitis
- innate immunity
1. Introduction
2. Clostridioides difficile Infection
“Bacillus difficile”, as denominated before, entered the scientific spotlight in 1935 when it was first identified by researchers Hall and O’Toole [10][9]. At that time, this bacterium was considered relatively innocuous, quietly inhabiting the human gut without drawing much attention. However, it was during the 1970s that “Clostridium difficile” began to reveal its pathogenic potential [11,12,13][10][11][12]. Researchers observed a connection between C. difficile and antibiotic-associated diarrhea, as well as pseudomembranous colitis, leading to a surge in research interest [8]. Since then, the bacterium has persisted in its evolutionary trajectory, developing resistance to various antibiotics and producing virulent strains that pose significant challenges to healthcare providers and researchers alike [14][13]. C. difficile is a Gram-positive bacterium characterized by its ability to form spores [16][14]. These spores are the resilient, dormant form of the bacterium, exhibiting the capacity to endure adverse conditions, including high temperatures and sterilizing chemicals [17][15]. Their durability contributes to their persistence in the environment, making CDI prevention and control challenging. Moreover, C. difficile spores play a critical role in CDI recurrence; even after successful CDI treatment, some patients experience a recurrence due to the germination of these spores when favorable conditions arise in the colon, often as a result of disruptions to the normal gut microbiota [16,18,19][14][16][17]. The pathological consequences of CDI are primarily centered in the colon. The disease can manifest across a spectrum of severity, ranging from mild diarrhea to severe pseudomembranous colitis, and in extreme cases, leading to fulminant outcomes [23,24,25][18][19][20]. Specifically, the development of pseudomembranous colitis arises as a sequence of unfolding events leading to colitis: CDI often starts with the ingestion of C. difficile spores, typically encountered in the environment, which then transform into active bacteria after germination within the large intestine (cecum and colon) [16][14]. C. difficile produces two crucial toxins, toxin A (TcdA) and toxin B (TcdB), that specifically target epithelial cells by binding to their respective receptors [5,6][5][6]. Once internalized, these toxins cause the subsequent inactivation of Rho-family GTPases through glucosylation [5,26,27][5][21][22]. This event disrupts the host cell’s cytoskeleton, accelerating cell death [28,29][23][24]. In addition, the emergence of hypervirulent strains, such as the NAP1/B1/027 variant, has emerged as a global public health concern by being associated with more severe and recurrent cases of CDI [33,34,35,36,37][25][26][27][28][29]. Specifically, the PCR ribotype 027 strain, commonly denoted as RT027, distinguishes itself with heightened virulence, characterized by elevated production of TcdA and TcdB toxins [38][30]. Infections with the RT027 strain are known for their high recurrence rates, impacting patient outcomes and placing a significant burden on healthcare systems. Remarkably, RT027 demonstrates resistance to certain antibiotics, notably fluoroquinolones, presenting challenges in treatment and contributing to persistent infections [39,40][31][32]. The current strategy for managing and treating patients with CDI typically involves a multifaceted approach [46][33]. Treatment plans are customized to address the unique clinical condition of each individual, underscoring the crucial role of early diagnosis and intervention for effectively managing the disease. The first step is discontinuing the antibiotic that may have triggered the infection, if applicable, to halt the selective advantage C. difficile gains in an altered gut microbiome. The next crucial step is the administration of specific antibiotics, such as vancomycin or fidaxomicin, to target and eliminate C. difficile bacteria [46,47,48][33][34][35]. Metronidazole is designated as an alternative agent and has been omitted from the treatment of nonsevere CDI since the 2021 guidelines [49][36]. Remarkably, fecal microbiota transplantation (FMT) surprisingly emerges as a highly potent therapeutic approach for treating recurrent or severe CDI [56,57][37][38]. This procedure entails the infusion of fecal matter from a healthy donor into the patient’s gastrointestinal tract, with the overarching goal of reestablishing a balanced gut microbiota. The transplanted microbiome, armed with its diverse arsenal of beneficial microbes, plays a crucial role in efficiently combatting the opportunistic C. difficile [3,58][3][39]. Meta-analyses examining the efficacy of FMT for recurrent, severe, or fulminant CDI consistently indicate promising outcomes [59,60][40][41]. FMT has demonstrated high success rates in preventing recurrent CDI, with significant reductions in recurrence compared to standard antibiotic therapy. Moreover, this therapeutic approach has shown effectiveness in treating severe and fulminant cases, contributing to improved clinical outcomes and reduced mortality rates. However, it is not without its drawbacks and potential risks [61][42]. One of the primary concerns is the lack of long-term safety data, as the consequences of introducing a new, diverse microbial community into a patient’s gut over time remain uncertain. FMT relies on donated fecal matter, which, despite rigorous donor screening and testing, may still carry undetected pathogens or infections that can be transmitted to the recipient [62,63][43][44]. There is also a risk of unintended alterations to the recipient’s microbiome, potentially leading to unexpected health issues [64,65][45][46]. Continuing research and ongoing debates focus on the application and long-term consequences of FMT, emphasizing the need for a thorough evaluation of its risks and benefits.3. Animal Models of CDI
CDI has been studied using different animal species, including hamsters, guinea pigs, rabbits, mice, and rats [9][47]. The hamster model is the most commonly used experimental model. On hamsters, CDI can be induced after ingestion of antibiotics, and colonization occurs through experimental challenge or environmental exposure to C. difficile [66,67,68][48][49][50]. The disease in this model primarily affects the cecum with some involvement of the ileum, resulting in diarrhea and fatal enterocolitis when exposed to toxigenic strains [69,70][51][52]. It is important to note that this model represents well the severe and lethal forms of the disease, but does not consistently display the full spectrum of CDI symptoms seen in humans [66,67,71][48][49][53]. Mice and rats are less susceptible to CDI than hamsters [73][54]. Nevertheless, murine models serve as valuable tools for studying CDI, given their genetic similarity to humans and enhanced translational relevance [74][55]. Mouse application allows researchers to control various factors such as genetics, environment, and diet, ensuring consistent experimental conditions. The availability of transgenic mice facilitates the examination of host factors and components of immune responses throughout the progression of the disease. There are well-established protocols for inducing C. difficile infection in mice, making them a standardized and widely adopted model in the field. Typically, laboratory mice are subjected to antibiotic treatment to disturb their gut microbiota, rendering them susceptible to the germination and colonization of C. difficile—mirroring the dynamics observed in humans [75,76][56][57]. Then, the mice are orally inoculated with C. difficile spores or vegetative cells. Over time, infected mice may develop symptoms similar to those seen in humans, such as diarrhea, weight loss, and other gastrointestinal issues [54,77][58][59]. The severity of symptoms varies depending on the C. difficile strain used and the susceptibility state of the mouse [78][60]. Numerous mouse models have been created for studying the mechanisms behind C. difficile pathogenesis, and each has its own strengths for investigating different aspects of the disease [75][56]. The selection of a particular method is guided by the research goals, the specific strain of C. difficile, and the intended disease features. One common approach is to perform a single oral gavage with C. difficile spores, as it mimics the natural route of infection from contaminated surfaces [80,81][61][62]. To study recurrent infections, some models involve multiple rounds of challenge [50,54][58][63]. Alternatively, researchers can administer purified C. difficile toxins to study the role of toxins in disease pathogenesis [82,83,84,85][64][65][66][67]. Antibiotic pretreatment disrupts the gut microbiota, making mice more susceptible to colonization, reflecting conditions seen in patients with antibiotic-associated C. difficile disease [86,87,88][68][69][70]. Gnotobiotic/germ-free mice can be colonized by C. difficile and exhibit intestinal pathology, primarily in the colon, with pseudomembrane formation similar to human disease [92,93,94,95][71][72][73][74]. This model in particular enables the exploration of microbiota or isolated groups of bacteria’s role in the development of CDI. Reeves et al. unveiled a significant delay in the onset of primary CDI and relapse in germ-free mice pretreated with probiotics [96][75]. Notably, the microbiome of mice undergoing moderate CDI and receiving probiotic treatment revealed a striking increase in the abundance of the Lachnospiraceae family during the initial CDI phase. Intriguingly, mice that were precolonized with the Lachnospiraceae isolate demonstrated a substantial decrease in C. difficile colonization, decreased levels of intestinal cytotoxin, and exhibited milder clinical symptoms and colonic histopathology. This effect was not observed in mice solely colonized with E. coli as a control. Thise study suggests novel therapeutic approaches for the treatment and prevention of CDI by utilizing bacterial species that potentially inhibit C. difficile growth. Likewise, a bacterial consortium named VE303 is currently under development as a potential treatment for high-risk CDI [97,98][76][77]. This oral treatment consists of eight nonpathogenic, nontoxigenic, commensal strains of Clostridia that have been shown to effectively restore a healthy gut microbial community, mitigate inflammation, and elevate levels of protective metabolites in mice [97,98][76][77]. However, the use of gnotobiotic mouse models has a much higher cost and is less practical when compared to conventional mice. Additionally, the absence of other microorganisms in these models makes them less representative of the human situation. Nonetheless, the utilization of germ-free mice is crucial for investigating the specific role of human microbiota in the development of CDI. Humanized mouse models, engrafted with human microbiota, provide an environment closer to human C. difficile infection [99,100,101,102][78][79][80][81].4. Antibiotic-Induced Murine Model of CDI
Antibiotic-induced murine models of CDI are specifically designed to mimic the clinical manifestations and pathological features of human CDI within a controlled laboratory setting [80,103][61][82]. Despite some limitations, this model has greatly contributed to our understanding of CDI and continues to be an essential component of CDI research. Antibiotics are employed to initiate gut dysbiosis, a crucial step in simulating the clinical conditions that lead to C. difficile expansion [87,104,105][69][83][84]. Commonly used antibiotics for this purpose include vancomycin, cefoperazone, and tetracycline, typically administered orally via drinking water supplementation [73,106,107,108,109][54][85][86][87][88]. The selection of antibiotics varies according to the specific study goals and the desired level of gut dysbiosis, with the administration of these antibiotics for a predetermined period before introducing C. difficile to initiate infection. Giel et al. conducted a study on the antibiotic-induced mouse model of CDI to investigate C. difficile spore germination [18][16]. Their findings revealed that spores were more likely to germinate in antibiotic-treated mice, but this germination was mitigated by cholestyramine, a treatment that chelated bile salts. This observation led the authors to suggest that cecal bacterial populations in antibiotic-treated mice had a reduced capacity to modify taurocholate, a spore germinant factor, providing further support for the role of bile salts in C. difficile spore germination and infection development in the host [18][16]. In some research scenarios, a combination of antibiotics may be utilized to induce a more profound disturbance in the gut microbiota, closely mirroring the clinical situation where patients often receive multiple antibiotics before developing CDI. Chen et al. [80][61] pioneered an innovative mouse model wherein C57Bl/6 mice were subjected to a 3-day oral exposure to a combination of kanamycin, gentamicin, colistin, metronidazole, and vancomycin, followed by an intraperitoneal administration of clindamycin 48 h later. Subsequently, the mice were challenged with varying doses of C. difficile. This mouse model faithfully replicated human C. difficile infections, inducing characteristic symptoms such as diarrhea, weight loss, and typical histological features. The severity of the disease in this mouse model was proportional to the challenge dose used, ranging from 2 × 102 to 105 CFU.5. CDI Pathogenesis and Disease Progression in Mice
After antibiotic treatment, the mouse’s gut undergoes substantial modifications, rendering it susceptible to C. difficile colonization [87,116,117][69][89][90]. The highly resilient C. difficile spores colonize the intestines, primarily finding their niche in the murine cecum and colon [16,118][14][91]. Within these regions, spores undergo germination, transitioning into vegetative cells that actively grow and produce the toxins responsible for clinical symptoms (Figure 1) [118][91]. Toxin A (TcdA) and toxin B (TcdB) are released following C. difficile germination, typically occurring on the first day postinfection [118][91]. These toxins have a specific affinity for the intestinal epithelial cells, disrupting the colonic epithelium and subsequently contributing to tissue damage and bacterial translocation [119,120,121,122][92][93][94][95]. In particular, gut bacterial translocation involves the process through which bacteria, mostly those that naturally inhabit the intestinal tract, cross the protective barrier of the gut and enter the bloodstream or other distant anatomical regions.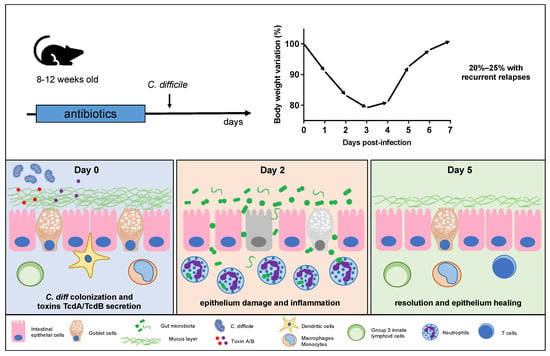
6. Conclusions
References
- Balsells, E.; Shi, T.; Leese, C.; Lyell, I.; Burrows, J.; Wiuff, C.; Campbell, H.; Kyaw, M.H.; Nair, H. Global Burden of Clostridium Difficile Infections: A Systematic Review and Meta-Analysis. J. Glob. Health 2019, 9, 010407.
- Bartlett, J.G.; Gerding, D.N. Clinical Recognition and Diagnosis of Clostridium Difficile Infection. Clin. Infect. Dis. 2008, 46, S12–S18.
- van Nood, E.; Vrieze, A.; Nieuwdorp, M.; Fuentes, S.; Zoetendal, E.G.; de Vos, W.M.; Visser, C.E.; Kuijper, E.J.; Bartelsman, J.F.W.M.; Tijssen, J.G.P.; et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium Difficile. N. Engl. J. Med. 2013, 368, 407–415.
- Czepiel, J.; Dróżdż, M.; Pituch, H.; Kuijper, E.J.; Perucki, W.; Mielimonka, A.; Goldman, S.; Wultańska, D.; Garlicki, A.; Biesiada, G. Clostridium Difficile Infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1211–1221.
- Kordus, S.L.; Thomas, A.K.; Lacy, D.B. Clostridioides Difficile Toxins: Mechanisms of Action and Antitoxin Therapeutics. Nat. Rev. Microbiol. 2022, 20, 285–298.
- Nibbering, B.; Gerding, D.N.; Kuijper, E.J.; Zwittink, R.D.; Smits, W.K. Host Immune Responses to Clostridioides Difficile: Toxins and Beyond. Front. Microbiol. 2021, 12, 804949.
- Tedesco, F.J. Clindamycin-Associated Colitis. Ann. Intern. Med. 1974, 81, 429.
- Bartlett, J.G.; Moon, N.; Chang, T.W.; Taylor, N.; Onderdonk, A.B. Role of Clostridium Difficile in Antibiotic-Associated Pseudomembranous Colitis. Gastroenterology 1978, 75, 778–782.
- Hall, I.C. INTESTINAL FLORA IN NEW-BORN INFANTS. Am. J. Dis. Child. 1935, 49, 390.
- George, W.L.; Sutter, V.L.; Goldstein, E.J.; Ludwig, S.L.; Finegold, S.M. Aetiology of Antimicrobial-Agent-Associated Colitis. Lancet 1978, 1, 802–803.
- Larson, H.E.; Price, A.B.; Honour, P.; Borriello, S.P. Clostridium Difficile and the Aetiology of Pseudomembranous Colitis. Lancet 1978, 1, 1063–1066.
- Bartlett, J.G. Clostridium Difficile: History of Its Role as an Enteric Pathogen and the Current State of Knowledge about the Organism. Clin. Infect. Dis. 1994, 18 (Suppl. S4), S265–S272.
- Markovska, R.; Dimitrov, G.; Gergova, R.; Boyanova, L. Clostridioides Difficile, a New “Superbug”. Microorganisms 2023, 11, 845.
- Shen, A. Clostridioides Difficile Spore Formation and Germination: New Insights and Opportunities for Intervention. Annu. Rev. Microbiol. 2020, 74, 545–566.
- Deakin, L.J.; Clare, S.; Fagan, R.P.; Dawson, L.F.; Pickard, D.J.; West, M.R.; Wren, B.W.; Fairweather, N.F.; Dougan, G.; Lawley, T.D. The Clostridium Difficile Spo0A Gene Is a Persistence and Transmission Factor. Infect. Immun. 2012, 80, 2704–2711.
- Giel, J.L.; Sorg, J.A.; Sonenshein, A.L.; Zhu, J. Metabolism of Bile Salts in Mice Influences Spore Germination in Clostridium Difficile. PLoS ONE 2010, 5, e8740.
- Sorg, J.A.; Sonenshein, A.L. Bile Salts and Glycine as Cogerminants for Clostridium Difficile Spores. J. Bacteriol. 2008, 190, 2505–2512.
- Cleary, R.K. Clostridium Difficile-Associated Diarrhea and Colitis: Clinical Manifestations, Diagnosis, and Treatment. Dis. Colon. Rectum 1998, 41, 1435–1449.
- Kelly, C.P.; Pothoulakis, C.; LaMont, J.T. Clostridium Difficile Colitis. N. Engl. J. Med. 1994, 330, 257–262.
- Mylonakis, E.; Ryan, E.T.; Calderwood, S.B. Clostridium Difficile--Associated Diarrhea: A Review. Arch. Intern. Med. 2001, 161, 525–533.
- Yang, Z.; Zhang, Y.; Huang, T.; Feng, H. Glucosyltransferase Activity of Clostridium Difficile Toxin B Is Essential for Disease Pathogenesis. Gut Microbes 2015, 6, 221–224.
- Bilverstone, T.W.; Garland, M.; Cave, R.J.; Kelly, M.L.; Tholen, M.; Bouley, D.M.; Kaye, P.; Minton, N.P.; Bogyo, M.; Kuehne, S.A.; et al. The Glucosyltransferase Activity of C. Difficile Toxin B Is Required for Disease Pathogenesis. PLoS Pathog. 2020, 16, e1008852.
- Chumbler, N.M.; Farrow, M.A.; Lapierre, L.A.; Franklin, J.L.; Haslam, D.B.; Goldenring, J.R.; Lacy, D.B. Clostridium Difficile Toxin B Causes Epithelial Cell Necrosis through an Autoprocessing-Independent Mechanism. PLoS Pathog. 2012, 8, e1003072.
- Kuehne, S.A.; Cartman, S.T.; Heap, J.T.; Kelly, M.L.; Cockayne, A.; Minton, N.P. The Role of Toxin A and Toxin B in Clostridium Difficile Infection. Nature 2010, 467, 711–713.
- Lim, S.K.; Stuart, R.L.; Mackin, K.E.; Carter, G.P.; Kotsanas, D.; Francis, M.J.; Easton, M.; Dimovski, K.; Elliott, B.; Riley, T.V.; et al. Emergence of a Ribotype 244 Strain of Clostridium Difficile Associated with Severe Disease and Related to the Epidemic Ribotype 027 Strain. Clin. Infect. Dis. 2014, 58, 1723–1730.
- Shin, B.-M.; Kuak, E.Y.; Yoo, S.J.; Shin, W.C.; Yoo, H.M. Emerging Toxin A-B+ Variant Strain of Clostridium Difficile Responsible for Pseudomembranous Colitis at a Tertiary Care Hospital in Korea. Diagn. Microbiol. Infect. Dis. 2008, 60, 333–337.
- Kim, J.; Kim, Y.; Pai, H. Clinical Characteristics and Treatment Outcomes of Clostridium Difficile Infections by PCR Ribotype 017 and 018 Strains. PLoS ONE 2016, 11, e0168849.
- López-Ureña, D.; Orozco-Aguilar, J.; Chaves-Madrigal, Y.; Ramírez-Mata, A.; Villalobos-Jimenez, A.; Ost, S.; Quesada-Gómez, C.; Rodríguez, C.; Papatheodorou, P.; Chaves-Olarte, E. Toxin B Variants from Clostridium Difficile Strains VPI 10463 and NAP1/027 Share Similar Substrate Profile and Cellular Intoxication Kinetics but Use Different Host Cell Entry Factors. Toxins 2019, 11, 348.
- Geric, B.; Carman, R.J.; Rupnik, M.; Genheimer, C.W.; Sambol, S.P.; Lyerly, D.M.; Gerding, D.N.; Johnson, S. Binary Toxin-Producing, Large Clostridial Toxin-Negative Clostridium Difficile Strains Are Enterotoxic but Do Not Cause Disease in Hamsters. J. Infect. Dis. 2006, 193, 1143–1150.
- Warny, M.; Pepin, J.; Fang, A.; Killgore, G.; Thompson, A.; Brazier, J.; Frost, E.; McDonald, L.C. Toxin Production by an Emerging Strain of Clostridium Difficile Associated with Outbreaks of Severe Disease in North America and Europe. Lancet 2005, 366, 1079–1084.
- Freeman, J.; Vernon, J.; Pilling, S.; Morris, K.; Nicholson, S.; Shearman, S.; Longshaw, C.; Wilcox, M.H. Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes Study Group The ClosER Study: Results from a Three-Year Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium Difficile Ribotypes, 2011–2014. Clin. Microbiol. Infect. 2018, 24, 724–731.
- Goldstein, E.J.C.; Citron, D.M.; Sears, P.; Babakhani, F.; Sambol, S.P.; Gerding, D.N. Comparative Susceptibilities to Fidaxomicin (OPT-80) of Isolates Collected at Baseline, Recurrence, and Failure from Patients in Two Phase III Trials of Fidaxomicin against Clostridium Difficile Infection. Antimicrob. Agents Chemother. 2011, 55, 5194–5199.
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium Difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, e1–e48.
- Stevens, V.W.; Nelson, R.E.; Schwab-Daugherty, E.M.; Khader, K.; Jones, M.M.; Brown, K.A.; Greene, T.; Croft, L.D.; Neuhauser, M.; Glassman, P.; et al. Comparative Effectiveness of Vancomycin and Metronidazole for the Prevention of Recurrence and Death in Patients with Clostridium Difficile Infection. JAMA Intern. Med. 2017, 177, 546–553.
- Louie, T.J.; Miller, M.A.; Mullane, K.M.; Weiss, K.; Lentnek, A.; Golan, Y.; Gorbach, S.; Sears, P.; Shue, Y.-K.; OPT-80-003 Clinical Study Group. Fidaxomicin versus Vancomycin for Clostridium Difficile Infection. N. Engl. J. Med. 2011, 364, 422–431.
- Johnson, S.; Lavergne, V.; Skinner, A.M.; Gonzales-Luna, A.J.; Garey, K.W.; Kelly, C.P.; Wilcox, M.H. Clinical Practice Guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 Focused Update Guidelines on Management of Clostridioides Difficile Infection in Adults. Clin. Infect. Dis. 2021, 73, e1029–e1044.
- Eiseman, B.; Silen, W.; Bascom, G.S.; Kauvar, A.J. Fecal Enema as an Adjunct in the Treatment of Pseudomembranous Enterocolitis. Surgery 1958, 44, 854–859.
- Smits, L.P.; Bouter, K.E.C.; de Vos, W.M.; Borody, T.J.; Nieuwdorp, M. Therapeutic Potential of Fecal Microbiota Transplantation. Gastroenterology 2013, 145, 946–953.
- Gupta, S.; Allen-Vercoe, E.; Petrof, E.O. Fecal Microbiota Transplantation: In Perspective. Therap Adv. Gastroenterol. 2016, 9, 229–239.
- Baunwall, S.M.D.; Lee, M.M.; Eriksen, M.K.; Mullish, B.H.; Marchesi, J.R.; Dahlerup, J.F.; Hvas, C.L. Faecal Microbiota Transplantation for Recurrent Clostridioides Difficile Infection: An Updated Systematic Review and Meta-Analysis. EClinicalMedicine 2020, 29–30, 100642.
- Song, Y.N.; Yang, D.Y.; Veldhuyzen van Zanten, S.; Wong, K.; McArthur, E.; Song, C.Z.; Ianiro, G.; Cammarota, G.; Kelly, C.; Fischer, M.; et al. Fecal Microbiota Transplantation for Severe or Fulminant Clostridioides Difficile Infection: Systematic Review and Meta-Analysis. J. Can. Assoc. Gastroenterol. 2022, 5, e1–e11.
- Sadowsky, M.J.; Khoruts, A. Faecal Microbiota Transplantation Is Promising but Not a Panacea. Nat. Microbiol. 2016, 1, 16015.
- Brandt, L.J.; Aroniadis, O.C. An Overview of Fecal Microbiota Transplantation: Techniques, Indications, and Outcomes. Gastrointest. Endosc. 2013, 78, 240–249.
- Kelly, B.J.; Tebas, P. Clinical Practice and Infrastructure Review of Fecal Microbiota Transplantation for Clostridium Difficile Infection. Chest 2018, 153, 266–277.
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214.
- Smith, M.I.; Yatsunenko, T.; Manary, M.J.; Trehan, I.; Mkakosya, R.; Cheng, J.; Kau, A.L.; Rich, S.S.; Concannon, P.; Mychaleckyj, J.C.; et al. Gut Microbiomes of Malawian Twin Pairs Discordant for Kwashiorkor. Science 2013, 339, 548–554.
- Best, E.L.; Freeman, J.; Wilcox, M.H. Models for the Study of Clostridium Difficile Infection. Gut Microbes 2012, 3, 145–167.
- Chang, T.W.; Bartlett, J.G.; Gorbach, S.L.; Onderdonk, A.B. Clindamycin-Induced Enterocolitis in Hamsters as a Model of Pseudomembranous Colitis in Patients. Infect. Immun. 1978, 20, 526–529.
- Toshniwal, R.; Silva, J.; Fekety, R.; Kim, K.H. Studies on the Epidemiology of Colitis Due to Clostridium Difficile in Hamsters. J. Infect. Dis. 1981, 143, 51–54.
- Toshniwal, R.; Fekety, R.; Silva, J. Etiology of Tetracycline-Associated Pseudomembranous Colitis in Hamsters. Antimicrob. Agents Chemother. 1979, 16, 167–170.
- Price, A.B.; Larson, H.E.; Crow, J. Morphology of Experimental Antibiotic-Associated Enterocolitis in the Hamster: A Model for Human Pseudomembranous Colitis and Antibiotic-Associated Diarrhoea. Gut 1979, 20, 467–475.
- Bartlett, J.G.; Onderdonk, A.B.; Cisneros, R.L.; Kasper, D.L. Clindamycin-Associated Colitis Due to a Toxin-Producing Species of Clostridium in Hamsters. J. Infect. Dis. 1977, 136, 701–705.
- Rifkin, G.D.; Silva, J.; Fekety, R. Gastrointestinal and Systemic Toxicity of Fecal Extracts from Hamsters with Clindamycin-Induced Colitis. Gastroenterology 1978, 74, 52–57.
- Lawley, T.D.; Clare, S.; Walker, A.W.; Goulding, D.; Stabler, R.A.; Croucher, N.; Mastroeni, P.; Scott, P.; Raisen, C.; Mottram, L.; et al. Antibiotic Treatment of Clostridium Difficile Carrier Mice Triggers a Supershedder State, Spore-Mediated Transmission, and Severe Disease in Immunocompromised Hosts. Infect. Immun. 2009, 77, 3661–3669.
- Collignon, A. Methods for Working with the Mouse Model. Methods Mol. Biol. 2010, 646, 229–237.
- Lawley, T.D.; Young, V.B. Murine Models to Study Clostridium Difficile Infection and Transmission. Anaerobe 2013, 24, 94–97.
- Britton, R.A.; Young, V.B. Interaction between the Intestinal Microbiota and Host in Clostridium Difficile Colonization Resistance. Trends Microbiol. 2012, 20, 313–319.
- Sun, X.; Wang, H.; Zhang, Y.; Chen, K.; Davis, B.; Feng, H. Mouse Relapse Model of Clostridium Difficile Infection. Infect. Immun. 2011, 79, 2856–2864.
- Li, Y.; Figler, R.A.; Kolling, G.; Bracken, T.C.; Rieger, J.; Stevenson, R.W.; Linden, J.; Guerrant, R.L.; Warren, C.A. Adenosine A2A Receptor Activation Reduces Recurrence and Mortality from Clostridium Difficile Infection in Mice following Vancomycin Treatment. BMC Infect. Dis. 2012, 12, 342.
- Castro-Córdova, P.; Díaz-Yáñez, F.; Muñoz-Miralles, J.; Gil, F.; Paredes-Sabja, D. Effect of Antibiotic to Induce Clostridioides Difficile-Susceptibility and Infectious Strain in a Mouse Model of Clostridioides Difficile Infection and Recurrence. Anaerobe 2020, 62, 102149.
- Chen, X.; Katchar, K.; Goldsmith, J.D.; Nanthakumar, N.; Cheknis, A.; Gerding, D.N.; Kelly, C.P. A Mouse Model of Clostridium Difficile-Associated Disease. Gastroenterology 2008, 135, 1984–1992.
- De Wolfe, T.J.; Kates, A.E.; Barko, L.; Darien, B.J.; Safdar, N. Modified Mouse Model of Clostridioides Difficile Infection as a Platform for Probiotic Efficacy Studies. Antimicrob. Agents Chemother. 2019, 63, e00111-19.
- Song, J.H.; Kim, Y.S. Recurrent Clostridium Difficile Infection: Risk Factors, Treatment, and Prevention. Gut Liver 2019, 13, 16–24.
- Lyerly, D.M.; Saum, K.E.; MacDonald, D.K.; Wilkins, T.D. Effects of Clostridium Difficile Toxins given Intragastrically to Animals. Infect. Immun. 1985, 47, 349–352.
- Hirota, S.A.; Iablokov, V.; Tulk, S.E.; Schenck, L.P.; Becker, H.; Nguyen, J.; Al Bashir, S.; Dingle, T.C.; Laing, A.; Liu, J.; et al. Intrarectal Instillation of Clostridium Difficile Toxin A Triggers Colonic Inflammation and Tissue Damage: Development of a Novel and Efficient Mouse Model of Clostridium Difficile Toxin Exposure. Infect. Immun. 2012, 80, 4474–4484.
- Savidge, T.C.; Pan, W.-H.; Newman, P.; O’brien, M.; Anton, P.M.; Pothoulakis, C. Clostridium Difficile Toxin B Is an Inflammatory Enterotoxin in Human Intestine. Gastroenterology 2003, 125, 413–420.
- Lucas, F.; Elmer, G.W.; Brot-Laroche, E.; Corthier, G. Fixation of Clostridium Difficile Toxin A and Cholera Toxin to Intestinal Brush Border Membranes from Axenic and Conventional Mice. Infect. Immun. 1989, 57, 1680–1683.
- Lesniak, N.A.; Schubert, A.M.; Flynn, K.J.; Leslie, J.L.; Sinani, H.; Bergin, I.L.; Young, V.B.; Schloss, P.D. The Gut Bacterial Community Potentiates Clostridioides Difficile Infection Severity. mBio 2022, 13, e0118322.
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-Induced Shifts in the Mouse Gut Microbiome and Metabolome Increase Susceptibility to Clostridium Difficile Infection. Nat. Commun. 2014, 5, 3114.
- Lawley, T.D.; Clare, S.; Walker, A.W.; Stares, M.D.; Connor, T.R.; Raisen, C.; Goulding, D.; Rad, R.; Schreiber, F.; Brandt, C.; et al. Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium Difficile Disease in Mice. PLoS Pathog. 2012, 8, e1002995.
- Pawlowski, S.W.; Calabrese, G.; Kolling, G.L.; Platts-Mills, J.; Freire, R.; AlcantaraWarren, C.; Liu, B.; Sartor, R.B.; Guerrant, R.L. Murine Model of Clostridium Difficile Infection with Aged Gnotobiotic C57BL/6 Mice and a BI/NAP1 Strain. J. Infect. Dis. 2010, 202, 1708–1712.
- Wilson, K.H.; Sheagren, J.N.; Freter, R.; Weatherbee, L.; Lyerly, D. Gnotobiotic Models for Study of the Microbial Ecology of Clostridium Difficile and Escherichia Coli. J. Infect. Dis. 1986, 153, 547–551.
- Onderdonk, A.B.; Cisneros, R.L.; Bartlett, J.G. Clostridium Difficile in Gnotobiotic Mice. Infect. Immun. 1980, 28, 277–282.
- Torres, J.; Jennische, E.; Lange, S.; Lönnroth, I. Enterotoxins from Clostridium Difficile; Diarrhoeogenic Potency and Morphological Effects in the Rat Intestine. Gut 1990, 31, 781–785.
- Reeves, A.E.; Koenigsknecht, M.J.; Bergin, I.L.; Young, V.B. Suppression of Clostridium Difficile in the Gastrointestinal Tracts of Germfree Mice Inoculated with a Murine Isolate from the Family Lachnospiraceae. Infect. Immun. 2012, 80, 3786–3794.
- Louie, T.; Golan, Y.; Khanna, S.; Bobilev, D.; Erpelding, N.; Fratazzi, C.; Carini, M.; Menon, R.; Ruisi, M.; Norman, J.M.; et al. VE303, a Defined Bacterial Consortium, for Prevention of Recurrent Clostridioides Difficile Infection: A Randomized Clinical Trial. JAMA 2023, 329, 1356–1366.
- Bobilev, D.; Bhattarai, S.; Menon, R.; Klein, B.; Reddy, S.; Olle, B.; Roberts, B.; Bucci, V.; Norman, J. 1953. VE303, a Rationally Designed Bacterial Consortium for Prevention of Recurrent Clostridioides Difficile (C. Difficile) Infection (RCDI), Stably Restores the Gut Microbiota After Vancomycin (Vanco)-Induced Dysbiosis in Adult Healthy Volunteers (HV). Open Forum Infect Dis 2019, 6, S60.
- Raibaud, P.; Ducluzeau, R.; Dubos, F.; Hudault, S.; Bewa, H.; Muller, M.C. Implantation of Bacteria from the Digestive Tract of Man and Various Animals into Gnotobiotic Mice. Am. J. Clin. Nutr. 1980, 33, 2440–2447.
- Hazenberg, M.P.; Bakker, M.; Verschoor-Burggraaf, A. Effects of the Human Intestinal Flora on Germ-Free Mice. J. Appl. Bacteriol. 1981, 50, 95–106.
- Péchiné, S.; Janoir, C.; Boureau, H.; Gleizes, A.; Tsapis, N.; Hoys, S.; Fattal, E.; Collignon, A. Diminished Intestinal Colonization by Clostridium Difficile and Immune Response in Mice after Mucosal Immunization with Surface Proteins of Clostridium Difficile. Vaccine 2007, 25, 3946–3954.
- Collins, J.; Auchtung, J.M.; Schaefer, L.; Eaton, K.A.; Britton, R.A. Humanized Microbiota Mice as a Model of Recurrent Clostridium Difficile Disease. Microbiome 2015, 3, 35.
- Schubert, A.M.; Sinani, H.; Schloss, P.D. Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium Difficile. mBio 2015, 6, e00974.
- Seekatz, A.M.; Safdar, N.; Khanna, S. The Role of the Gut Microbiome in Colonization Resistance and Recurrent Clostridioides Difficile Infection. Therap. Adv. Gastroenterol. 2022, 15, 17562848221134396.
- Reeves, A.E.; Theriot, C.M.; Bergin, I.L.; Huffnagle, G.B.; Schloss, P.D.; Young, V.B. The Interplay between Microbiome Dynamics and Pathogen Dynamics in a Murine Model of Clostridium Difficile Infection. Gut Microbes 2011, 2, 145–158.
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound Alterations of Intestinal Microbiota following a Single Dose of Clindamycin Results in Sustained Susceptibility to Clostridium Difficile-Induced Colitis. Infect. Immun. 2012, 80, 62–73.
- Theriot, C.M.; Koumpouras, C.C.; Carlson, P.E.; Bergin, I.I.; Aronoff, D.M.; Young, V.B. Cefoperazone-Treated Mice as an Experimental Platform to Assess Differential Virulence of Clostridium Difficile Strains. Gut Microbes 2011, 2, 326–334.
- Theriot, C.M.; Schumacher, C.A.; Bassis, C.M.; Seekatz, A.M.; Young, V.B. Effects of Tigecycline and Vancomycin Administration on Established Clostridium Difficile Infection. Antimicrob. Agents Chemother. 2015, 59, 1596–1604.
- Kiełbasiński, K.; Peszek, W.; Grabarek, B.O.; Boroń, D.; Wierzbik-Strońska, M.; Oplawski, M. Effect of Salinomycin on Expression Pattern of Genes Associated with Apoptosis in Endometrial Cancer Cell Line. Curr. Pharm. Biotechnol. 2020, 21, 1269–1277.
- Brown, K.A.; Khanafer, N.; Daneman, N.; Fisman, D.N. Meta-Analysis of Antibiotics and the Risk of Community-Associated Clostridium Difficile Infection. Antimicrob. Agents Chemother. 2013, 57, 2326–2332.
- Yurist-Doutsch, S.; Arrieta, M.-C.; Vogt, S.L.; Finlay, B.B. Gastrointestinal Microbiota-Mediated Control of Enteric Pathogens. Annu. Rev. Genet. 2014, 48, 361–382.
- Koenigsknecht, M.J.; Theriot, C.M.; Bergin, I.L.; Schumacher, C.A.; Schloss, P.D.; Young, V.B. Dynamics and Establishment of Clostridium Difficile Infection in the Murine Gastrointestinal Tract. Infect. Immun. 2015, 83, 934–941.
- Feltis, B.A.; Kim, A.S.; Kinneberg, K.M.; Lyerly, D.L.; Wilkins, T.D.; Erlandsen, S.L.; Wells, C.L. Clostridium Difficile Toxins May Augment Bacterial Penetration of Intestinal Epithelium. Arch. Surg. 1999, 134, 1235–1241.
- Naaber, P.; Mikelsaar, R.H.; Salminen, S.; Mikelsaar, M. Bacterial Translocation, Intestinal Microflora and Morphological Changes of Intestinal Mucosa in Experimental Models of Clostridium Difficile Infection. J. Med. Microbiol. 1998, 47, 591–598.
- Fachi, J.L.; Sécca, C.; Rodrigues, P.B.; de Mato, F.C.P.; Di Luccia, B.; Felipe, J.d.S.; Pral, L.P.; Rungue, M.; Rocha, V.D.M.; Sato, F.T.; et al. Acetate Coordinates Neutrophil and ILC3 Responses against C. Difficile through FFAR2. J. Exp. Med. 2020, 217, e20190489.
- Hasegawa, M.; Yamazaki, T.; Kamada, N.; Tawaratsumida, K.; Kim, Y.-G.; Núñez, G.; Inohara, N. Nucleotide-Binding Oligomerization Domain 1 Mediates Recognition of Clostridium Difficile and Induces Neutrophil Recruitment and Protection against the Pathogen. J. Immunol. 2011, 186, 4872–4880.
- Hasegawa, M.; Kamada, N.; Jiao, Y.; Liu, M.Z.; Núñez, G.; Inohara, N. Protective Role of Commensals against Clostridium Difficile Infection via an IL-1β-Mediated Positive-Feedback Loop. J. Immunol. 2012, 189, 3085–3091.
- Rocha, M.F.; Maia, M.E.; Bezerra, L.R.; Lyerly, D.M.; Guerrant, R.L.; Ribeiro, R.A.; Lima, A.A. Clostridium Difficile Toxin A Induces the Release of Neutrophil Chemotactic Factors from Rat Peritoneal Macrophages: Role of Interleukin-1beta, Tumor Necrosis Factor Alpha, and Leukotrienes. Infect. Immun. 1997, 65, 2740–2746.
- Abt, M.C.; Lewis, B.B.; Caballero, S.; Xiong, H.; Carter, R.A.; Sušac, B.; Ling, L.; Leiner, I.; Pamer, E.G. Innate Immune Defenses Mediated by Two ILC Subsets Are Critical for Protection against Acute Clostridium Difficile Infection. Cell Host Microbe 2015, 18, 27–37.
- Jarchum, I.; Liu, M.; Shi, C.; Equinda, M.; Pamer, E.G. Critical Role for MyD88-Mediated Neutrophil Recruitment during Clostridium Difficile Colitis. Infect. Immun. 2012, 80, 2989–2996.
- McDermott, A.J.; Falkowski, N.R.; McDonald, R.A.; Frank, C.R.; Pandit, C.R.; Young, V.B.; Huffnagle, G.B. Role of Interferon-γ and Inflammatory Monocytes in Driving Colonic Inflammation during Acute Clostridium Difficile Infection in Mice. Immunology 2017, 150, 468–477.
- Cowardin, C.A.; Kuehne, S.A.; Buonomo, E.L.; Marie, C.S.; Minton, N.P.; Petri, W.A. Inflammasome Activation Contributes to Interleukin-23 Production in Response to Clostridium Difficile. mBio 2015, 6, e02386-14.
- Ng, J.; Hirota, S.A.; Gross, O.; Li, Y.; Ulke-Lemee, A.; Potentier, M.S.; Schenck, L.P.; Vilaysane, A.; Seamone, M.E.; Feng, H.; et al. Clostridium Difficile Toxin-Induced Inflammation and Intestinal Injury Are Mediated by the Inflammasome. Gastroenterology 2010, 139, 542–552.
