Entomopathogenic bacteria and fungi are quite frequently found in soils and insect cadavers. The first step in utilizing these microbes as biopesticides is to isolate them, and several culture media and insect baiting procedures have been tested in this direction.
- Beauveria
- Metarhizium
- Hypocreales
- Bacillus thuringiensis
- Serratia
1. Introduction
|
Bacteria |
Target Pest |
Crops |
PRODUCT (Company, Country) |
|---|---|---|---|
|
Bacillus thuringiensis subsp. kurstaki |
Lepidoptera |
Row crops, forests, orchards, forests turfs |
CRYMAX (Certis, USA) |
|
DELIVER (Certis, USA) |
|||
|
JAVELIN WG (Certis, USA) |
|||
|
COSTAR JARDIN; COSTAR WG (Mitsui AgriScience International NV, Belgium) |
|||
|
LEPINOX PLUS (CBC, Europe) |
|||
|
BACTOSPEINE JARDIN EC (Duphar BV, The Netherlands) |
|||
Pastures | |||
BIOSHIELD GRASS GRUB (Biostart, New Zealand) | |||
Paenibacillus popilliae | |||
Japanese beetle larvae/grub | |||
Lawns, flowers, mulch beds, gardens |
MILKY SPORE POWDER (St. Gabriel Organics, USA) |
|
Fungi |
Target Pest |
Crop |
Product and Company |
||||
|---|---|---|---|---|---|---|---|
|
Beauveria bassiana sensu lato |
Psyllids, whiteflies, thrips, aphids, mites |
crops |
BOTE GHA (Certis, USA) |
||||
|
Flies, mites, thrips, leafhoppers, and weevils |
cotton, glasshouse crops |
NATURALIS (Troy Biosciences, USA) |
|||||
|
Coffee berry borer |
coffee |
CONIDIA (AgroEvo, Germany) |
|||||
|
Whiteflies, aphids, thrips |
field crops |
MYCOTROL (Bioworks, USA) |
|||||
|
Whiteflies, aphids, thrips |
field crops |
BOTANIGRAD (Bioworks, USA) |
|||||
|
Corn borer |
maize |
OSTRINIL (Arysta Lifescience, France) |
|||||
|
DOLPHIN (Andermatt Biocontrol, Switzerland) |
|||||||
|
Spotted mite, eucalyptus weevil, coffee borer, and whitefly |
crops |
BOVERIL (Koppert, The Netherlands) |
BMP 123 (Becker, USA) |
||||
|
Flies |
BALANCE (Rincon-Vitova Insectaries, USA) |
DIPEL DF (Valent Biosciences, USA) |
|||||
|
As soil treatment |
crops |
BEAUVERIA BASSIANA PLUS, (BuildASoil, USA) |
LEAP (Valent Biosciences, USA) |
||||
|
Whitefly |
peppers, tomatoes, potatoes, eggplants |
BEA-SIN (Agrobionsa, Mexico) |
FORAY 48 B (Valent Biosciences, USA) |
||||
|
B. brongniartii |
May beetle |
forests, vegetables, fruits, grasslands |
MELOCONT PILZGERSTE (Samen-schwarzenberger, Austria) |
B. thuringiensis subsp. aizawai |
Lepidoptera |
Row crops, orchards |
CRYMAX (Certis, USA) |
|
Cockchafer larvae |
Fruits, Meadows |
BEAUPRO (Andermatt Biocontrol, Switzerland) |
AGREE 50 WG (Certis, USA) |
||||
|
Scarabs beetle larvae |
sugarcane |
BETEL (Natural Plant Protection, France) |
XENTARI (Valent Biosciences, USA) |
||||
|
Cockchafer |
fruits, Meadows |
BEAUVERIA-SCHWEIZER (Eric Schweizer, Switzerland) |
FLORBAC (Bayer, Germany) |
||||
|
Metarhizium anisopliae sensu lato |
Sugar cane root leafhopper |
sugarcane |
METARRIL WP (Koppert, The Netherlands) |
B. thuringiensis subsp. tenebrionis |
|||
|
Cockroaches |
Coleoptera: Chrysomelidae |
houses | Potatoes, tomatoes, eggplant, elm trees |
BIO-PATH (EcoScience, USA) TRIDENT (Certis USA) |
|||
|
NOVODOR FC (Valent Biosciences, USA) |
|||||||
|
Vine weevils, sciarid flies, wireworms and thrips pupae |
glasshouse, ornamental crops |
BIO 1020 (Bayer, Germany) |
B. thuringiensis subsp. israelensis |
||||
|
White grubs | Diptera |
sugarcane |
Diverse lentic and lotic aquatic habitats |
AQUABAC DF3000, (Becker Microbial Products Inc, USA) |
|||
BIOCANE (BASF, Australia) |
VECTOPRIME (Valent Biosciences, USA) |
||||||
|
termites |
BIOBLAST (Paragon, USA) |
TEKNAR (Valent Biosciences, USA) |
|||||
|
Black vine weevil, strawberry root weevil, thrips |
stored grains and crops |
MET-52 (Novozymes, USA) |
VECTOBAC (Valent Biosciences, USA) |
||||
|
Pepper weevil |
chili and bell peppers |
META-SIN (Agrobionsa, Mexico) |
BACTIMOS (Valent Biosciences, USA) |
||||
|
M. acridum |
Locusts and grasshoppers |
crops |
GREEN GUARD (BASF, Australia) |
SOLBAC (Andermatt Biocontrol, Switzerland) |
|||
|
M. frigidum |
Scarab larvae |
crops |
BIOGREEN (BASF, Australia) |
Lysinibacillus sphaericus |
Diptera: Culicidae |
||
|
M. brunneum |
Wireworms |
Lentic aquatic habitats |
VECTOLEX (Valent Biosciences, USA) |
||||
potato and asparagus crops |
ATTRACAP (Biocare, Germany) |
Serratia entomophila |
Coleoptera: Scarabaeidae | ||||
|
Cordyceps fumosorosea |
Whiteflies |
glasshouse crops |
PREFERAL WG (Biobest, Belgium) |
||||
|
Aphids, Citrus psyllid, spider mite, thrips, whitefly |
wide range of crops |
PFR-97 20% WDG (Certis, USA) |
|||||
|
Whitefly |
Peppers, tomatoes, potatoes, eggplants |
BEA-SIN (Agrobionsa, Mexico) |
|||||
|
Cotton bullworm, Citrus psyllid |
Field crops |
CHALLENGER (Koppert, The Netherlands) |
|||||
|
Lecanicillium longisporum |
Aphids |
crops |
VERTALEC (Koppert, The Netherlands) |
||||
|
Whiteflies, thrips |
crops |
MYCOTAL (Koppert, The Netherlands) |
|||||
|
L. lecanii |
Aphids |
peppers, tomatoes, potatoes, eggplants |
VERTI-SIN (Agrobionsa, Mexico) |
2. Isolation of Entomopathogenic Bacteria
2.1. Milky Disease-Causing Paenibacillus spp.
- (a)
-
Disinfect the surface of the larvae of grubs (Coleoptera) with 0.5% (v/v) sodium hypochlorite (NaOCl).
- (b)
- (a)
-
Make soil suspensions by adding 2 g soil to 20 mL sterilized water.
- (b)
-
Make a germinating medium, i.e., 0.5% yeast extract and 0.1% glucose.
- (c)
-
Adjust the pH to 6.5.
- (d)
-
Add germinating medium into the soil suspension at 1:50 ratio.
- (e)
-
Apply series of heat shocks at 70 °C for 20 min after every hour, 7 times.
- (f)
-
Spread the aliquot on J-Medium and incubate for 7 h at 28 °C, anaerobically.
2.2. Amber Disease-Causing Serratia spp.
- (a)
-
Soil inoculums or hemolymph of the diseased larvae can be isolated on Caprylate-thallous agar (CTA) [31][25].
- (b)
-
Culturing is done by pulling and separating the anterior end of the cadavers. The gut contents are then cultured on CTA plates.
- (c)
-
Serratia marcescens produces colonies which are red in color. Cream-colured bacterial colonies formed on CTA can then be transferred into different selective media for the identification of Serratia spp. [30][24].
- (d)
-
The production of a halo on a Deoxyribonuclease (DNase)-Toluidine Blue agar when incubated at 30 °C for 24 h, indicates the presence of Serratia spp. [32][26]. Thereafter, the production of blue or green colonies on adonitol agar confirms S. proteamaculans. The formation of yellow colonies on adonitol agar hints the presence of S. entomophila, which can be confirmed by the growth on itaconate agar at 30 °C after 96 h [25][19]. Further molecular approaches targeting specific DNA regions can distinguish pathogenic strains from the non-pathogenic ones.
2.3. Other Bacteria from the Class Bacilli
- (a)
-
Isolation can be done from soils (2–4 g in 10 mL sterilized water), insects (0.2–0.4 g/mL sterilized water), or water samples (after concentrating using 0.22 µm filter).
- (b)
-
Heat the samples in a water bath at 80 °C for 10 min to kill the vegetative cells.
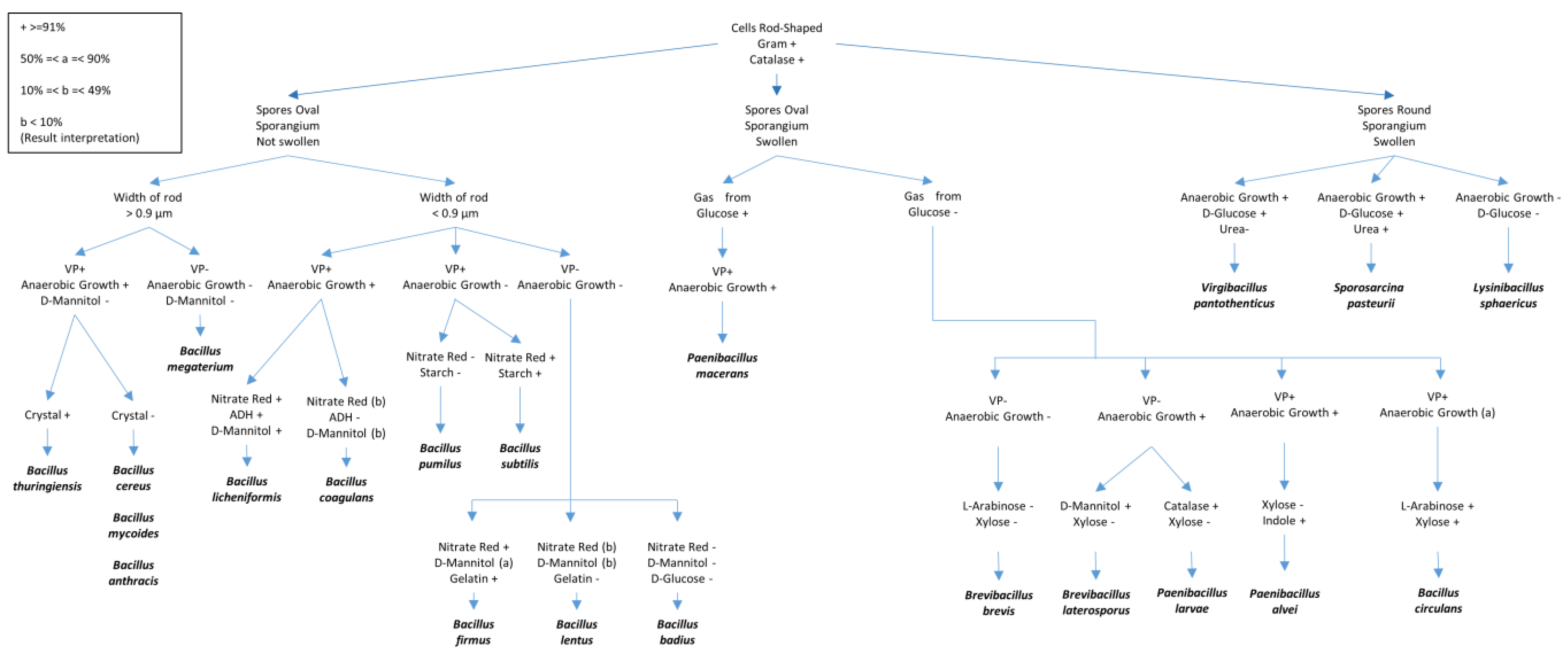
- Figure 1. Different biochemical tests for the identification of Bacilli species. The figure was adapted and redrawn after modifications from T.W. Fisher and Garczynski [23]. Some details of the tests presented include VP (Voges–Proskauer test (Barritt’s method)), Gelatin (proteolysis of gelatin), ADH (presence of the amino acid arginine dihydrolase), Glucose (fermentation) and Mannitol (fermentation); Starch (hydrolysis), Nitrate (nitrate reduction to nitrite), and Urea (Urease test).
Different biochemical tests for the identification of Bacilli species. The figure was adapted and redrawn after modifications from T.W. Fisher and Garczynski [17]. Some details of the tests presented include VP (Voges–Proskauer test (Barritt’s method)), Gelatin (proteolysis of gelatin), ADH (presence of the amino acid arginine dihydrolase), Glucose (fermentation) and Mannitol (fermentation); Starch (hydrolysis), Nitrate (nitrate reduction to nitrite), and Urea (Urease test).
3. Isolation of Entomopathogenic Fungi
3.1. Isolations from Naturally Mycosed Insect Cadavers
- (a)
-
Insect cadavers are brought to the laboratory as separate entities in sterile tubes.
- (b)
-
Insects are observed under a stereomicroscope (40×) for probable mycosis.
- (c)
-
In case of a visible mycosis, the insects are surface sterilized using 70% ethanol or 1% NaOCl, for 3 min, followed by 3 distinct washes with 100 mL of sterilized water. Then, the sporulating EPF from the insect cadaver is plated directly.
- (d)
-
Cadavers are then cultured on a selective medium at 22 °C for up to 3 weeks, depending on the time taken by the fungi for germination and proliferation. In case of no germination, the cadavers can be homogenized and plated on the selective medium. Details of the different selective medium are provided later in the text.
- (e)
-
Obtained fungi are subcultured on potato dextrose agar (PDA) or Sabouraud dextrose agar (SDA) until pure culture is obtained.
3.2. Isolations from Soils
3.3. Isolation from Phyllosphere
Some studies have also isolated EPF from the phylloplane and other parts of the plant phyllosphere, as these fungi can also be present as plant epiphytes or endophytes [41][35]. Meyling et al. suggested a leaf imprinting methodology where the leaf is cultured onto a selective agar medium [64][43]. Petri dishes with partitions are used and the upper (adaxial), and the lower (abaxial) surface of the leaf are pressed on the separate sides of the petri plate. Henceforth, the same leaf is put on a paper sheet and photocopied to estimate its surface area using image analysis software at a later stage. The petri plates are incubated in the dark at 23 °C to count fungal colony forming units (CFUs) [64][43]. Surface sterilization is quite important in isolating hypocrealean fungi as endophytes. This can be done by dipping the plant part in either 70% ethanol and/or 1–5% NaOCl for 3 min. In case of the leaves, the petiole can be first kept out of the sanitizer to avoid the chemical reaching inside the leaf, and then it can be cut to culture the sterilized part of the leaf on either of the selective mediums described above. It is always recommended to sanitize the intact plant part and then cut it into pieces for further culturing, as this avoids the sterilization of the endophytic fungi [111][44]. Different studies have isolated EPF from the phyllosphere, such as bark and branch samples [56,112][45][46] and leaves [59,113][47][48].3.4. Molecular Identifications of the Isolated Entomopathogenic Fungi
After obtaining a single spore fungal culture on a PDA or SDA, the species can be resolved or identified by amplifying the regions of nuclear ribosomal DNA, such as nrITS, large (28S) subunit (nrLSU), or small (18S) subunit (nrSSU). Another, nuclear ribosomal DNA region, i.e., the intergenic spacer region between nrSSU and nrLSU or IGS, has also been used to understand Beauveria and Metarhizium speciation [113,114,115,116][48][49][50][51]. The resolution of the molecular identification can be increased by amplifying other nuclear DNA regions of interest, e.g., for Bloc for Beauveria [113,114,115][48][49][50] and the 5′ intron-containing region of translation elongation factor 1-alpha subunit (5′-tef1α) for Metarhizium [116,117][51][52]. Other nuclear DNA markers, such as the regions of the gene encoding for the largest subunit of RNA polymerase II (rpb1), the second largest subunit of RNA polymerase II (rpb2); β-tublin (β-tub), and the coding region of Tef1-α, can also be employed, in general, for any EPF [118,119][53][54]. Moreover, in the last decades, researchers have been constantly developing and validating the use of several microsatellite markers for the genotyping of Beauveria [93,115,120,121,122,123][50][55][56][57][58][59] and Metarhizium [124,125][60][61] isolates. For example, Oulevey et al. [125][61] described 18 small single repeats or microsatellite marker sets for Metarhizium, i.e., Ma145, Ma325, Ma307, Ma2049, Ma2054, Ma2055, Ma2056, Ma2057, Ma2060, Ma2063, Ma2069, Ma2070, Ma2077, Ma2089, Ma2283, Ma2287, Ma2292, and Ma2296. Similarly, Meyling et al. [93][55] and Goble et al. [123][59] validated the use of 17 to 18 microsatellite marker sets for Beauveria, i.e., Ba06, Ba08, and Ba12-Ba29. This methodology enables enhanced resolution among very closely related isolates which may otherwise be rendered as clones. Recently, Kepler and Rehner [119][54] developed primers for the amplification and sequencing of nuclear intergenic spacer markers for the resolution of Metarhizium isolates, i.e., BTIGS, MzFG543, MzFG546, MzIGS2, MzIGS3, MzIGS5, and MzIGS7, and Kepler et al. [99][62] successfully validated the use of MzIGS3 and MzFG543 on the Metarhizium isolated from agricultural soils.References
- Ruiu, L. Microbial Biopesticides in Agroecosystems. Agronomy 2018, 8, 235.
- Glare, T.; Caradus, J.; Gelernter, W.; Jackson, T.; Keyhani, N.; Köhl, J.; Marrone, P.; Morin, L.; Stewart, A. Have biopesticides come of age? Trends Biotechnol. 2012, 30, 250–258.
- Marx-Stoelting, P.; Pfeil, R.; Solecki, R.; Ulbrich, B.; Grote, K.; Ritz, V.; Banasiak, U.; Heinrich-Hirsch, B.; Moeller, T.; Chahoud, I.; et al. Assessment strategies and decision criteria for pesticides with endocrine disrupting properties relevant to humans. Reprod. Toxicol. 2011, 31, 574–584.
- Sharma, L.; Gonçalves, F.; Oliveira, I.; Torres, L.; Marques, G. Insect-associated fungi from naturally mycosed vine mealybug Planococcus ficus (Signoret) (Hemiptera: Pseudococcidae). Biocontrol Sci. Technol. 2018, 28, 122–141.
- Sharma, L.; Marques, G. Fusarium, an Entomopathogen—A Myth or Reality? Pathogens 2018, 7, 93.
- Sharma, L.; Oliveira, I.; Raimundo, F.; Torres, L.; Marques, G. Soil chemical properties barely perturb the abundance of entomopathogenic Fusarium oxysporum: A case study using a generalized linear mixed model for microbial pathogen occurrence count data. Pathogens 2018, 7, 89.
- Sharma, L.; Oliveira, I.; Torres, L.; Marques, G. Entomopathogenic fungi in Portuguese vineyards soils: Suggesting a ‘Galleria-Tenebrio-bait method’ as bait-insects Galleria and Tenebrio significantly underestimate the respective recoveries of Metarhizium (robertsii) and Beauveria (bassiana). MycoKeys 2018, 38, 1–23.
- Sharma, L.; Bohra, N.; Singh, R.K.; Marques, G. Potential of Entomopathogenic Bacteria and Fungi. In Microbes for Sustainable Insect Pest Management: An Eco-friendly Approach—Volume 1; Khan, M.A., Ahmad, W., Eds.; Springer: Cham, Switzerland, 2019; pp. 115–149.
- Azizoglu, U.; Jouzani, G.S.; Yilmaz, N.; Baz, E.; Ozkok, D. Genetically modified entomopathogenic bacteria, recent developments, benefits and impacts: A review. Sci. Total Environ. 2020, 734, 139169.
- Faria, M.R.d.; Wraight, S.P. Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control. 2007, 43, 237–256.
- Clifton, E.H.; Jaronski, S.T.; Hajek, A.E. Virulence of commercialized fungal entomopathogens against asian longhorned beetle (Coleoptera: Cerambycidae). J. Insect Sci. 2020, 20, 1.
- Ruiu, L. Insect Pathogenic Bacteria in Integrated Pest Management. Insects 2015, 6, 352.
- Godjo, A.; Afouda, L.; Baimey, H.; Decraemer, W.; Willems, A. Molecular diversity of Photorhabdus and Xenorhabdus bacteria, symbionts of Heterorhabditis and Steinernema nematodes retrieved from soil in Benin. Arch. Microbiol. 2018, 200, 589–601.
- Hurst, M.R.H.; Becher, S.A.; Young, S.D.; Nelson, T.L.; Glare, T.R. Yersinia entomophaga sp. nov., isolated from the New Zealand grass grub Costelytra zealandica. Int. J. Syst. Evol. Microbiol. 2011, 61, 844–849.
- Vodovar, N.; Vinals, M.; Liehl, P.; Basset, A.; Degrouard, J.; Spellman, P.; Boccard, F.; Lemaitre, B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. USA 2005, 102, 11414–11419.
- Martin, P.A.W.; Hirose, E.; Aldrich, J.R. Toxicity of Chromobacterium subtsugae to southern green stink bug (Heteroptera: Pentatomidae) and corn rootworm (Coleoptera: Chrysomelidae). J. Econ. Entomol. 2007, 100, 680–684.
- Stahly, D.P.; Takefman, D.M.; Livasy, C.A.; Dingman, D.W. Selective medium for quantitation of Bacillus popilliae; in soil and in commercial spore powders. Appl. Environ. Microbiol. 1992, 58, 740.
- Stahly, D.P.; Andrews, R.E.; Yousten, A.A. The genus Bacillus—Insect pathogens. In The Prokaryotes: Volume 4: Bacteria: Firmicutes, Cyanobacteria; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 563–608.
- Koppenhöfer, A.M.; Jackson, T.; Klein, M.G. Bacteria for Use Against Soil-Inhabiting Insects; Academic Press: San Diego, CA, USA, 2012; pp. 129–149.
- St. Julian, G.J.; Pridham, T.G.; Hall, H.H. Effect of diluents on viability of Popillia japonica Newman larvae, Bacillus popilliae Dutky, and Bacillus lentimorbus Dutky. J. Invertebr. Pathol. 1963, 5, 440–450.
- Dingman, D.W.; Stahly, D.P. Medium Promoting Sporulation of Bacillus larvae and Metabolism of Medium Components. Appl. Environ. Microbiol. 1983, 46, 860–869.
- Krieger, L.; Franken, E.; Schnetter, W. Bacillus popilliae var melolontha H1, a pathogen for the May beetles, Melolontha spp. In Proceedings of the 3rd International Workshop on Microbial Control of Soil Dwelling Pests, Lincoln, New Zealand, 21–23 February 1996; Jackson, T.A., Glare, T.R., Eds.; AgResearch: Lincoln, New Zealand, 1996; pp. 79–87.
- Milner, R.J. A method for isolating milky disease, Bacillus popilliae var. rhopaea, spores from the soil. J. Invertebr. Pathol. 1977, 30, 283–287.
- O’Callaghan, M.; Jackson, T.A. Isolation and enumeration of Serratia entomophila—a bacterial pathogen of the New Zealand grass grub, Costelytra zealandica. J. Appl. Bacteriol. 1993, 75, 307–314.
- Starr, M.P.; Grimont, P.A.; Grimont, F.; Starr, P.B. Caprylate-thallous agar medium for selectively isolating Serratia and its utility in the clinical laboratory. J. Clin. Microbiol. 1976, 4, 270.
- Berkowitz, D.M.; Lee, W.S. A selective medium for the isolation and identification of Serratia marcescens. In Abstracts of the Annual Meeting of the American Society for Microbiology; American Society for Microbiology: Washington, DC, USA, 1973; Volume 105.
- Kalfon, A.; Larget-Thiéry, I.; Charles, J.-F.; de Barjac, H. Growth, sporulation and larvicidal activity of Bacillus sphaericus. Eur. J. Appl. Microbiol. Biotechnol. 1983, 18, 168–173.
- Fisher, T.W.; Garczynski, S.F. Chapter III—Isolation, culture, preservation, and identification of entomopathogenic bacteria of the Bacilli. In Manual of Techniques in Invertebrate Pathology, 2nd ed.; Lacey, L.A., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 75–99.
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as Biological Control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41.
- Meza-Menchaca, T.; Singh, R.K.; Quiroz-Chávez, J.; García-Pérez, L.M.; Rodríguez-Mora, N.; Soto-Luna, M.; Gastélum-Contreras, G.; Vanzzini-Zago, V.; Sharma, L.; Quiroz-Figueroa, F.R. First demonstration of clinical Fusarium strains causing cross-kingdom infections from humans to plants. Microorganisms 2020, 8, 947.
- Carlos, C.G.F.; Sousa, S.; Salvação, J.; Sharma, L.; Soares, R.; Manso, J.; Nóbrega, M.; Lopes, A.; Soares, S.; Aranha, J.; et al. Environmentally safe strategies to control the European Grapevine Moth, Lobesia botrana (Den. & Schiff.) in the Douro Demarcated Region. Cienc. Tec. Vitivinic. 2013, 28, 1006–1011.
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi, 2nd ed.; IHW-Verlag and Verlagsbuchhandlung: Eching, Germany, 2007.
- Humber, R.A. Chapter VI—Identification of entomopathogenic fungi. In Manual of Techniques in Invertebrate Pathology, 2nd Ed.; Lacey, L.A., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 151–187.
- Yurkov, A.; Guerreiro, M.A.; Sharma, L.; Carvalho, C.; Fonseca, Á. Correction: Multigene assessment of the species boundaries and sexual status of the basidiomycetous yeasts Cryptococcus flavescens and C. terrestris (Tremellales). PLoS ONE 2015, 10, e0126996.
- Inglis, G.D.; Enkerli, J.; Goettel, M.S. Chapter VII—Laboratory techniques used for entomopathogenic fungi: Hypocreales. In Manual of Techniques in Invertebrate Pathology, 2nd ed.; Lacey, L.A., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 189–253.
- Ramos, Y.; Portal, O.; Lysøe, E.; Meyling, N.V.; Klingen, I. Diversity and abundance of Beauveria bassiana in soils, stink bugs and plant tissues of common bean from organic and conventional fields. J. Invertebr. Pathol. 2017, 150, 114–120.
- Sun, B.-D.; Liu, X.-Z. Occurrence and diversity of insect-associated fungi in natural soils in China. Appl. Soil Ecol. 2008, 39, 100–108.
- Oliveira, I.; Pereira, J.A.; Lino-Neto, T.; Bento, A.; Baptista, P. Fungal diversity associated to the olive moth, Prays oleae Bernard: A survey for potential entomopathogenic fungi. Microb. Ecol. 2012, 63, 964–974.
- Oliveira, I.; Pereira, J.A.; Quesada-Moraga, E.; Lino-Neto, T.; Bento, A.; Baptista, P. Effect of soil tillage on natural occurrence of fungal entomopathogens associated to Prays oleae Bern. Sci. Hortic. 2013, 159, 190–196.
- Greenfield, M.; Gómez-Jiménez, M.I.; Ortiz, V.; Vega, F.E.; Kramer, M.; Parsa, S. Beauveria bassiana and Metarhizium anisopliae endophytically colonize cassava roots following soil drench inoculation. Biol. Control 2016, 95, 40–48.
- Posadas, J.B.; Comerio, R.M.; Mini, J.I.; Nussenbaum, A.L.; Lecuona, R.E. A novel dodine-free selective medium based on the use of cetyl trimethyl ammonium bromide (CTAB) to isolate Beauveria bassiana, Metarhizium anisopliae sensu lato and Paecilomyces lilacinus from soil. Mycologia 2012, 104, 974–980.
- King, A.D.; Hocking, A.D.; Pitt, J.I. Dichloran-rose bengal medium for enumeration and isolation of molds from foods. Appl. Environ. Microbiol. 1979, 37, 959–964.
- Meyling, N.V.; Eilenberg, J. Isolation and characterisation of Beauveria bassiana isolates from phylloplanes of hedgerow vegetation. Mycol. Res. 2006, 110, 188–195.
- Ownley, B.H.; Griffin, M.R.; Klingeman, W.E.; Gwinn, K.D.; Moulton, J.K.; Pereira, R.M. Beauveria bassiana: Endophytic colonization and plant disease control. J. Invertebr. Pathol. 2008, 98, 267–270.
- Ormond, E.L.; Thomas, A.P.; Pugh, P.J.; Pell, J.K.; Roy, H.E. A fungal pathogen in time and space: The population dynamics of Beauveria bassiana in a conifer forest. FEMS Microbiol. Ecol. 2010, 74, 146–154.
- Nishi, O.; Sushida, H.; Higashi, Y.; Iida, Y. Epiphytic and endophytic colonisation of tomato plants by the entomopathogenic fungus Beauveria bassiana strain GHA. Mycology 2020, 1–9.
- Clifton, E.H.; Jaronski, S.T.; Coates, B.S.; Hodgson, E.W.; Gassmann, A.J. Effects of endophytic entomopathogenic fungi on soybean aphid and identification of Metarhizium isolates from agricultural fields. PLoS ONE 2018, 13, e0194815.
- Meyling, N.V.; Pilz, C.; Keller, S.; Widmer, F.; Enkerli, J. Diversity of Beauveria spp. isolates from pollen beetles Meligethes aeneus in Switzerland. J. Invertebr. Pathol. 2012, 109, 76–82.
- Rehner, S.A.; Posada, F.; Buckley, E.P.; Infante, F.; Castillo, A.; Vega, F.E. Phylogenetic origins of African and neotropical Beauveria bassiana s.l. pathogens of the coffee berry borer, Hypothenemus hampei. J. Invertebr. Pathol. 2006, 93, 11–21.
- Meyling, N.V.; Lubeck, M.; Buckley, E.P.; Eilenberg, J.; Rehner, S.A. Community composition, host range and genetic structure of the fungal entomopathogen Beauveria in adjoining agricultural and seminatural habitats. Mol. Ecol. 2009, 18, 1282–1293.
- Bischoff, J.F.; Rehner, S.A.; Humber, R.A. A multilocus phylogeny of the Metarhizium anisopliae lineage. Mycologia 2009, 101, 512–530.
- Rezende, J.M.; Zanardo, A.B.R.; da Silva Lopes, M.; Delalibera, I.; Rehner, S.A. Phylogenetic diversity of Brazilian Metarhizium associated with sugarcane agriculture. BioControl 2015, 60, 495–505.
- Spatafora, J.W.; Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; White, J.F. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 2007, 16, 1701–1711.
- Kepler, R.M.; Rehner, S.A. Genome-assisted development of nuclear intergenic sequence markers for entomopathogenic fungi of the Metarhizium anisopliae species complex. Mol. Ecol. Resour. 2013, 13, 210–217.
- Steinwender, B.M.; Enkerli, J.; Widmer, F.; Eilenberg, J.; Thorup-Kristensen, K.; Meyling, N.V. Molecular diversity of the entomopathogenic fungal Metarhizium community within an agroecosystem. J. Invertebr. Pathol. 2014, 123, 6–12.
- Enkerli, J.; Widmer, F.; Gessler, C.; Keller, S. Strain-specific microsatellite markers in the entomopathogenic fungus Beauveria brongniartii. Mycol. Res. 2001, 105, 1079–1087.
- Rehner, S.A.; Buckley, E.P. Isolation and characterization of microsatellite loci from the entomopathogenic fungus Beauveria bassiana (Ascomycota: Hypocreales). Mol. Ecol. Notes 2003, 3, 409–411.
- Enkerli, J.; Widmer, F. Molecular ecology of fungal entomopathogens: Molecular genetic tools and their applications in population and fate studies. BioControl 2010, 55, 17–37.
- Goble, T.A.; Costet, L.; Robene, I.; Nibouche, S.; Rutherford, R.S.; Conlong, D.E.; Hill, M.P. Beauveria brongniartii on white grubs attacking sugarcane in South Africa. J. Invertebr. Pathol. 2012, 111, 225–236.
- Enkerli, J.; Kölliker, R.; Keller, S.; Widmer, F. Isolation and characterization of microsatellite markers from the entomopathogenic fungus Metarhizium anisopliae. Mol. Ecol. Notes 2005, 5, 384–386.
- Oulevey, C.; Widmer, F.; Kölliker, R.; Enkerli, J. An optimized microsatellite marker set for detection of Metarhizium anisopliae genotype diversity on field and regional scales. Mycol. Res. 2009, 113, 1016–1024.
- Kepler, R.M.; Ugine, T.A.; Maul, J.E.; Cavigelli, M.A.; Rehner, S.A. Community composition and population genetics of insect pathogenic fungi in the genus Metarhizium from soils of a long-term agricultural research system. Environ. Microbiol. 2015, 17, 2791–2804.