Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Mariana Romo-Castillo.
Klebsiella pneumoniae (Kpn) is an opportunistic pathogen that causes intrahospital complications such as pneumonia, liver abscesses, soft tissue infections, urinary infections, bacteraemia, and, in some cases, death. Since this bacterium has a higher frequency than other Gram-negative pathogens, it has become an important pathogen to the health sector. The adaptative genome of Kpn likely facilitates increased survival of the pathogen in diverse situations.
- Klebsiella pneumoniae
- virulence factors
- antibiotic resistance
- plant-origin compounds
1. Introduction: General Aspects of Klebsiella pneumoniae
Klebsiella pneumoniae (Kpn) was first isolated by Carl Friedlander in 1882 from the lungs of dead pneumonia patients [1]. Hence, it was initially called Friedlander’s bacillus, but in 1886, it was renamed Kpn. This Gram-negative bacillus, an encapsulated, non-motile bacterium, is a lactose-fermenting, non-spore-forming facultative anaerobe that belongs to the Enterobacteriaceae family (Figure 1) [2].
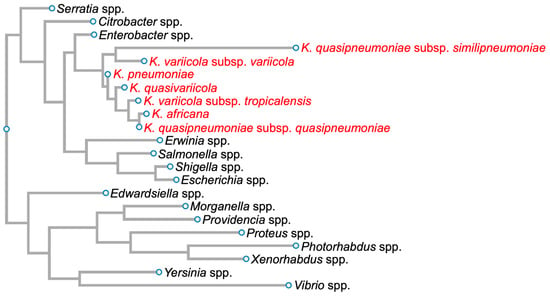
Figure 1. Phylogenetic tree of members of the family Enterobacteriaceae based on 16S rDNA sequences. Members of the Klebsiella pneumoniae species complex are in red. Diagram was generated for graphical information display using 16S rDNA sequences of Serratia marcescens strain Gol3 (ID: MT263018.1), Citrobacter freundii strain S12 (ID: MW879533.1), Enterobacter cloacae strain ATCC 13047 1 (ID: NR_102794.2), Klebsiella quasipneumoniae subsp. similipneumoniae strain CW-D 3 (ID: NR_132596.1), Klebsiella variicola strain 13450 (ID: CP026013.1), Klebsiella pneumoniae strain DSM 30104 (ID: NR_036794.1), Klebsiella quasivariicola strain KPN1705 (ID: OQ719747.1), Klebsiella variicola subsp. tropicalensis strain VITGAJ4 (ID: MT829337.1), Klebsiella africana strain SB5857 (ID: MK040622.1), Klebsiella quasipneumoniae subsp. quasipneumoniae strain 01A030 (ID: NR_134062.1), Erwinia amylovora strain S34 (ID: OP512541.1), Salmonella enterica strain 16OCT84 (ID: OQ581800.1), Shigella dysenteriae strain ATCC 13313 (ID: NR_026332.1), Escherichia coli strain Gol11 16S (ID: MT263026.1), Edwardsiella tarda strain KC-Pc-HB1(ID: CP023706.1), Morganella morganii subsp. morganii strain 229813 (ID: CP043955.1), Providencia stuartii strain ATCC 29914 (ID: NR_024848.1), Proteus vulgaris ATCC 29905 (ID: NR_115878.1), Photorhabdus asymbiotica strain ATCC 43951 (ID: Z76754.1), Xenorhabdus nematophila strain DSM3370 (ID: NR_119150.1), Yersinia pestis strain SCPM-O-B-6291 (ID: CP045163.1), and Vibrio cholerae strain RC782(ID: ON849168.1).
Kpn is a Klebsiella pneumoniae species complex (KpSC) member, which includes strains that have the same biochemical profile but only share 90–96% identity between their genome sequences [3,4,5][3][4][5]. This complex contains seven phylogroups: Klebsiella pneumoniae, Klebsiella quasipneumoniae subsp. quasipneumoniae, Klebsiella quasipneumoniae subsp. similipneumoniae, Klebsiella variicola subsp. variicola, Klebsiella variicola subsp. tropicalensis, Klebsiella quasivariicola, and Klebsiella africana (Figure 1).
Although all these strains could be associated with human diseases, Kpn is mainly related to nosocomial infections [5]. Kpn strains are classified into three categories: classical (cKpn), hypervirulent (hvKpn), and multidrug-resistant (MDR-Kpn). cKpn strains are nosocomial strains that are generally found in immunocompromised patients and produce urinary tract, acute respiratory, and bloodstream infections [5]. cKpn strains are characterised by a non-hypermucoviscosity phenotype (“string test”) and lack excessive siderophores [6]. hvKpn strains are community-acquired strains that induce pyogenic liver abscess, meningitis, endophthalmitis, and necrotising fasciitis in diabetic and healthy people. These strains are characterised by a hypermucoviscosity phenotype and harbour excessive siderophores [7]. Finally, pyogenic liver abscesses, bloodstream infections, and urinary tract infections are associated with nosocomial and community MDR-Kpn strains that usually infect immunocompromised patients [8]. MDR-Kpn strains have a hypervirulent profile but no specific hypermucoviscosity-defined pattern [6,9][6][9]. MDR-Kpn and hvKpn strains were previously considered different clonal groups, but now these characteristics are deemed additive [10]. The high incidence of MDR-Kpn and hvKpn strains has earned Kpn a place on the World Health Organization (WHO) list as a “priority pathogen” since 2017 [11].
The complex biology of Kpn has made it difficult to control infections caused by this pathogen due to its high antimicrobial resistance and many virulence factors. That is why designing new therapies to combat Kpn infections has been a priority. Plants are a potential source for the design of new treatments [12]. Numerous biotic and abiotic stimuli, including certain pathogenic bacteria, cause plants to produce many secondary metabolites. These metabolites are employed in medicine to treat many illnesses, including infections involving bacteria. However, very few studies have demonstrated these metabolites’ antimicrobial potential, mechanism of action, and potential for developing new antibacterial therapies.
2. Kpn Virulence Factors
Kpn has acquired many virulence factors to promote colonisation, evasion of host immune responses, and bacterial competition (Figure 2).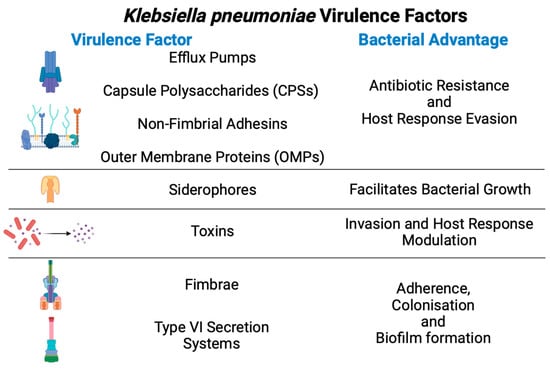
Figure 2.
General virulence factors from
Klebsiella pneumoniae
and their advantages during infection.
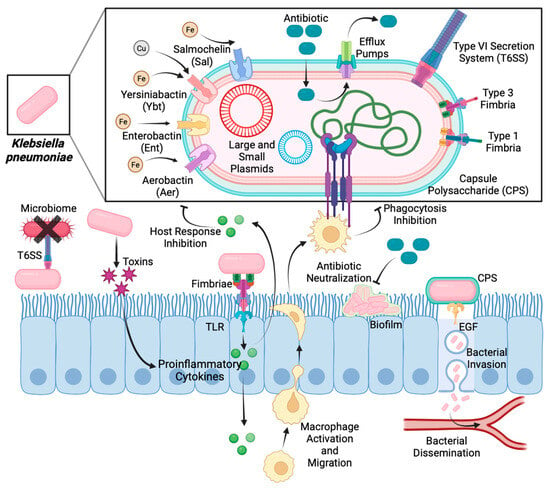
Figure 3.
Mechanisms of action of virulence factors during
Klebsiella pneumoniae
infection.
3. Klebsiella pneumoniae and Public Health
Kpn is the most frequent nosocomial opportunistic pathogen that infects critically ill and immunocompromised patients. This pathogen has become a severe public health problem due to its high prevalence and mortality rate in hospitals [35]. Many reports describing Kpn disease have focused on adults. Nevertheless, recent studies indicate that children (5.9–67.6%) are susceptible to infection by hvKpn strains [36,37][36][37]. As previously mentioned, the diseases produced by Kpn include bacteraemia, liver diseases, pneumonia, urinary tract infections, and septic arthritis [38,39,40][38][39][40]. Furthermore, it is one of the most important opportunistic pathogens to combat during solid organ transplants [41,42][41][42]. Haematological malignancies, antibiotic administration, mechanical ventilation, and long-term hospitalisations are the principal factors that favour Kpn dissemination in hospitalised patients [43,44,45][43][44][45]. Interestingly, the transmission of Kpn disease during the SARS-CoV-2 pandemic was significantly reduced, along with the transmission of Enterococcus faecium, Staphylococcus aureus, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. (called ESKAPE pathogens) [46,47][46][47]. Government-implemented public health measures, such as using face masks and gloves, meticulous hand cleaning, access and movement restrictions within the hospitals, and continuous surface cleaning, could be causes of the low incidence of ESKAPE pathogens [48]. Even though antibiotic therapy is ineffective against SARS-CoV-2 infection, some studies reported that 70% of hospitalised patients received broad-spectrum antibiotic therapy [49]. Furthermore, a recent mathematical-method-based study analysed the impact of health interventions on the prevalence of resistant Kpn strains; in that study, an increase in extensively drug-resistant strains was reported, with the frequency rising from 10% to 50% [50]. Genomic studies and sequence type (ST) analysis have suggested that ST307 and ST147 strains have a high number of virulence factors (antibacterial resistance genes, virulence factors, and fitness); these factors are considered a potential risk because they may contribute to the adaptation of these strains to hospital environments and the human host [51]. Kpn ST307 was described in the Netherlands in 2008; however, after five years, the clone was also reported in hospitals in the United States, Pakistan, Colombia, Italy, South Korea, and Tunisia [52]. After 2016, Kpn ST307 was dispersed worldwide, likely as a result of acquiring a second capsule cluster, a fimbrial cluster, and T6SS [53]. The Kpn ST147 clone was first reported in 2008–2009 in Hungary and Spain and is mainly resistant to the fluoroquinolones due to gyrA S83I, parC S80IQRDR, and blaCTX-M-15 mutations [54]. From 2011 to 2013, the clone was identified in Greece, Italy, Sweden, Denmark, Canada, the United Kingdom, Finland, India, and Libya. Finally, in 2014, this clone achieved worldwide dispersion [52]. The impact of this uncontrolled antibiotic administration was a subsequent increase in resistant bacterial infections, highlighting the importance of tighter control in prophylactic practice and antibiotic administration. Thus, increasing efforts are being made to develop innovative, specific, and effective treatments to combat the severe and often deadly infections produced by Kpn strains.4. Antibiotic Resistance
Antibiotic resistance results from bacterial specialisation across time and exposure to different drugs as a bacterial survival strategy [55,56,57][55][56][57]. The main antibiotic-resistance-associated mechanisms are active efflux, target protection, decreased influx, target modification, LPS modification, physical barrier, and antibiotic inactivation (Figure 4).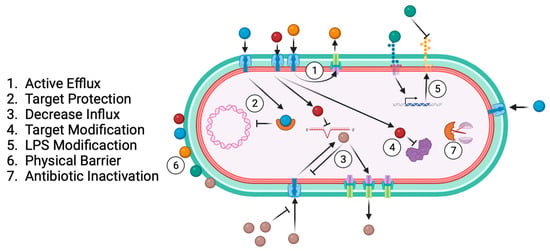
Figure 4.
Common bacterial antibiotic resistance mechanisms.
Table 1.
Principal mechanisms and genes related to antibiotic resistance in
Klebsiella pneumoniae
.
| Antibiotic | Resistance Mechanisms | Related Genes | Reference | ||
|---|---|---|---|---|---|
| β-Lactams | Antibiotic inactivation | blaSHV-2 | [61] | ||
| blaTEM-3 | [62] | ||||
| blaCTX-M | [63] | ||||
| blaAMP | [64] | ||||
| Acetone leaf extract | MIC = 128 µg/mL | [ | 111] | blaIMP-1 | [65] |
| Acacia nilotica | Aqueous extract | MIC = 11.7 mg/mL MBC = 13.3 mg/mL Reduces biofilm by 59.03% |
[112] | ||
| Himatanthus drasticus | Hydroalcoholic extract | Zone diameter = 16 ± 0.5 mm MIC and MBC = 6250 µg/mL Reduces biofilm by 50% |
7. Biotechnology Applying Plant-Origin Sources to Develop New Control Strategies against Kpn
An important factor in controlling and reducing nosocomial infections, such as Kpn disease, is the implementation of new technologies that control intrahospital dispersion of the strains. Many studies have reported that hospital personnel, the environment, and medical devices are the principal routes by which these opportunistic pathogens are transferred to patients [141]. To solve this problem, nanotechnology researchers are working on a nanoparticle (NP) design that could be useful in bionic, textile, and biomedical engineering (Figure 6).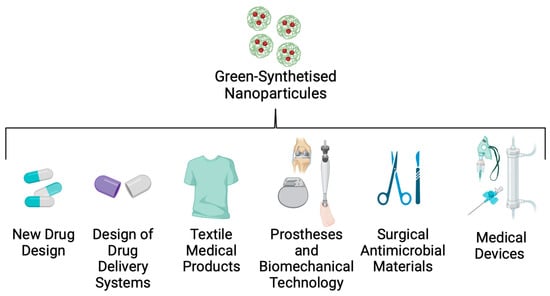
Figure 6.
Applications of green-synthesised nanoparticles in biomedicine.
8. Special Considerations in the Application of POCs in Medicine
It is important to note that POCs are a complex mixture of secondary metabolites produced by plants, and their chemical composition can change from plant to plant, even within the same species, as a result of exposure to biotic and abiotic factors such as soil hydrology, pH, salinity, temperature, soil organisms, and pollinator insects. Additionally, postharvest treatment, extraction, and conservation methods can influence POCs’ chemical composition. These results have been the subject of numerous examinations, such as the study performed by Todorova et al. in 2023 [152]. They analysed the chemical composition of commercial and bio-cultivated lavender (Lavandula angustifolia Mill.) essential oil. While bio-cultivated lavender essential oil contains 23.13% β-linalool, commercial essential oils possess higher amounts (24.34–35.99%). Additionally, only one of the seven samples contained less than 25% linalyl acetate, the content indicated by the requirements of the European Pharmacopoeia. The pharmacological potential of POCs depends on their composition. Establishing them as an official drug could be difficult, and to solve the trouble of composition variations, standardisation of plant growth, postharvest, and extraction protocols is required. While employing POCs can be among the most effective approaches for developing novel treatments, it is crucial to acknowledge that numerous secondary metabolites synthesised by plants serve as defence mechanisms against infections, insects, and herbivores. These mechanisms function by causing harm to organisms that consume them. Consequently, evaluating the cytotoxic potential of these metabolites is imperative to guarantee their suitability for medicinal plant applications. Although POCs are frequently commended for their aromatic and therapeutic benefits, it is vital to recognise that they can also carry hazards, including the possibility of poisoning. For example, essential oils are highly concentrated plant extracts that can have adverse effects due to the intensity or high concentration of some of their compounds. Recently, many studies have been carried out to characterize this cytotoxicity in vitro and in vivo. Table 3 describes the safety and toxicity data of many POCs that are proposed to have positive effects against Kpn.Table 3. Toxicity and safety assessment of some plant-origin compounds that are effective in combating Klebsiella pneumoniae infections.
| Plant | Model | Activity | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Momordica charantia | In vitro in lymphocytes | Lymphocyte viability was 98% at 12.5, 25, and 50 µg/mL), and micronucleus frequency was the same as in the negative control. M. charantia extracts did not affect IL-6 or IL-10 production. | [108] | |||||||
| In vivo in Wistar rats | The acute toxicity test revealed the manifestation of toxic signs in response to the hydroalcoholic extract of M. charantia, attributed to the presence of ethanol in the extract. A marginal reduction in body weight, although statistically nonsignificant, was observed. Conversely, administering the aqueous extract did not induce toxic signs or mortality. Both extracts were categorised as class 5, indicating their placement in the toxicity range with an LD50 greater than 2000 mg/kg. In the dermal and ocular irritation test, both extracts were deemed non-irritant. | [153] | ||||||||
| Bacopa monnieri | In vivo in Sprague–Dawley rats | B. monnieri extract (5000 mg/kg) did not cause a histopathological change in the internal organs, including the liver and the kidneys. Rats treated with B. monnieri extract at 30, 60, 300, and 1500 mg/kg dosages for 270 days did not present any toxic effect. |
[154] | |||||||
| Paeonia officinalis | In vivo in Wistar rats | Aqueous extracts of the roots of P. officinalis in an acute oral toxicity test did not cause mortality in rats at a dose of 175 mg/kg, 550 mg/kg, or 2000 mg/kg and were considered safe. | [155] | |||||||
| Acacia nilotica | In vitro in freshly collected human red blood cells | A. nilotica at doses of 50 μg/mL, 100 μg/mL, 150 μg/mL, and 200 μg/mL were found to possess haemolytic activity. | [156] | |||||||
| blaKPC | [66] | |||||||||
| Himatanthus drasticus | In vitro in human erythrocytes and peripheral blood mononuclear cells (PBMCs) | Himatanthus drasticus hydroalcoholic extract did not produce significant haemolysis at the concentrations tested, and no significant changes were detected in viability or nitric oxide (NO) production by PBMCs. | [113] | |||||||
| [ | 113 | ] | blaGEN | [67] | ||||||
| blaOXA-48 | [68] | |||||||||
| blaVIM-1 | [69] | |||||||||
| Permeability alterations | blaACT-1 | [70] | ||||||||
| [71] | ||||||||||
| Pulicaria crispa | Polyphenolic extract | Zone diameter values vary between 12.55 ± 0.31 and 24.00 ± 0.02 mm. MIC values range from 0.1 to 0.425 mg/mL |
[114] | |||||||
| Symplocos racemosa | Aminoglycosides | Drug modification | aac | [72] | ||||||
| ant | ||||||||||
| Ethyl acetate extract | aph | |||||||||
| Target protection | armA | [73] | ||||||||
| rmt | [72] | |||||||||
| npmA | ||||||||||
| Permeability alterations | ||||||||||
| Symplocos racemosa | In vivo using Swiss albino mice | In vivo acute oral toxicity testing did not show any toxic effects | [115kpnEF | [59] | ||||||
| ] | Zone diameter ranges from 14.33 to 25.66 mm | Quinolones | Target modification | gyrA 1 | [74] | |||||
| MIC ranges from 0.5 to 10.0 mg/mL | [ | 115 | ] | gyrB 1 | [75] | |||||
| parC 1 | [76] | |||||||||
| parE 1 | ||||||||||
| Origanum vulgare | In vivo using mice | Continuous use or high doses may deliver undesirable components causing liver and renal function impairment | [157] | Vaccinium corymbosum | Polyphenolic extract | Reduces the number of attached bacteria and biofilm production by 90% in vitro at 430 µg/mL | [116] | Permeability alterations | Plasmid-mediated quinolone resistance genes (PMQR) | [77] |
| Target protection | qnr | [78] | ||||||||
| Drug modification | aac(6′)-Ib-cr | [79] | ||||||||
| Polymyxin | LPS modification | LPS-MS 2 | [80] | |||||||
| lpxM | [81] | |||||||||
| pbgP | [82] | |||||||||
| pmrE | [83] | |||||||||
| mcr-1 | [84] | |||||||||
| pmrC | [85] | |||||||||
| pagP | ||||||||||
| phoPQ | ||||||||||
| pmrA | ||||||||||
| pmrD | ||||||||||
| Physical barrier | CPSs 3 | [86] | ||||||||
| Tigecycline | Permeability alterations | rarA | [87] | |||||||
| ramA | [88] | |||||||||
| ramR | [89] | |||||||||
| acrR | ||||||||||
| rpsJ | ||||||||||
| kpgA | ||||||||||
| kpgB | ||||||||||
| kpgC |
1 Mutations in these genes are responsible for antibiotic resistance; the mere presence of the genes is not. 2 LPS-MS = lipopolysaccharide modification system, 3 CPSs = capsule polysaccharides.
Sequestering antibiotic molecules inside the bacteria is another resistance mechanism for preventing attachment to their target; it is called “targeting protection”. This mechanism involves the physical association of a target protection protein, and the interaction does not cause a permanent change in the target or the antibiotic [90]. Quinolones form a complex with type II topoisomerases to inhibit the synthesis of bacterial DNA, providing resistance to infection. However, the Qnr pentapeptide repeat protein adopts a structure mimicking B-form DNA and binding to topoisomerase [91]. Kpn harbouring a qnr plasmid exhibited the MDR phenotype [92]. In addition, target modification is another mechanism of antibiotic resistance. Mutation of DNA gyrase (gyrA-gyrB subunits) or subunits of topoisomerase IV (parC-parE) is the principal cause of quinolone resistance [74]. A mutation of DNA gyrase (commonly in gyrB) is responsible for nalidixic acid resistance [56]. Modifying the lipopolysaccharide (LPS) structure is a mechanism bacteria develop to affect the target [93]. In the presence of some antibiotics, such as polymyxin, bacteria activate a lipopolysaccharide modification system that involves altering the LPS maturation proteins and incorporating new chains. The Kpn genes responsible for this phenotype are lpxM, pmrE, and pafP, which are involved in lipid maturation [81,82,83][81][82][83].
Antibiotic inactivation is an enzyme-based process where bacterial enzymes modify antibiotic activity by hydrolysis or redox modification, beta-lactamase being the most representative example of this mechanism [57].
Many hydrophilic drugs use porin channels to penetrate bacterial membranes. The regulation of efflux-pump expression is a strategy some bacteria use to limit penetration by these drugs [57]. Efflux pumps such as KdeA, AcrAB, and OqxAB are essential for controlling cell permeability and are involved in quinolone resistance [77,94,95][77][94][95].
Additionally, physical barriers such as capsules and biofilms affect antibiotic diffusion and provide the pathogen with resistance [96]. The antibiotic resistance mechanisms of Kpn highlight the relevance of developing new therapies that could complement or even substitute for antibiotic treatments.
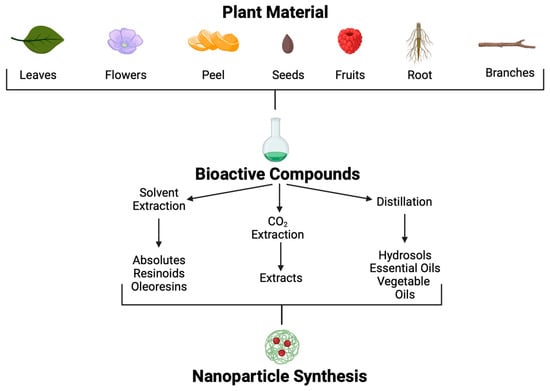 Different extraction methods are used to obtain these POCs [107]. Through solvent extraction, absolutes, resinoids, and oleoresins can be obtained. Absolutes are aromatic liquids extracted by maceration with chemical solvents, such as ethanol or hexane, followed by filtration and concentration to produce wax. Resinoids are obtained by chemical extraction with solvents from extracts of resinous plant exudates. In contrast, oleoresins are obtained from spice extraction with a hydrocarbon solvent followed by vacuum distillation. On the other hand, aromatic polar and non-polar compounds found in plants at low concentrations, such as essential oils and hydrosols, can be obtained by distillation.
Many plant extracts with antibacterial activity against Kpn strains have been developed (Table 2). For example, ref. [107], Momordica charantia leaf ethanolic extract was also efficient against cKpn strains [108], and Skimmia anquetilia root extract and Bacopa monnieri methanolic and ethanolic leaf extracts were effective against MDR-Kpn strains [109,110][109][110].
Different extraction methods are used to obtain these POCs [107]. Through solvent extraction, absolutes, resinoids, and oleoresins can be obtained. Absolutes are aromatic liquids extracted by maceration with chemical solvents, such as ethanol or hexane, followed by filtration and concentration to produce wax. Resinoids are obtained by chemical extraction with solvents from extracts of resinous plant exudates. In contrast, oleoresins are obtained from spice extraction with a hydrocarbon solvent followed by vacuum distillation. On the other hand, aromatic polar and non-polar compounds found in plants at low concentrations, such as essential oils and hydrosols, can be obtained by distillation.
Many plant extracts with antibacterial activity against Kpn strains have been developed (Table 2). For example, ref. [107], Momordica charantia leaf ethanolic extract was also efficient against cKpn strains [108], and Skimmia anquetilia root extract and Bacopa monnieri methanolic and ethanolic leaf extracts were effective against MDR-Kpn strains [109,110][109][110].
Moreover, Paeonia officinalis showed bacteriostatic activity against MDR-Kpn strains but only when water subfractionation was used, indicating the importance of selecting accurate extraction solvents and subfractionation methods in the search for effective POCs with antibacterial potential [111].
Biofilm formation represents the major challenge in treating Kpn disease, since the construction of this structure restricts penetration by any molecule [129]. Many plant-based products have demonstrated high antibiofilm activity against Kpn, such as Acacia nilotica aqueous extract [112], Himatanthus drasticus hydroalcoholic extract [113], Pulicaria crispa polyphenolic extract [114], Symplocos racemosa ethyl acetate extract [115], Vaccinium corymbosum water extract [116], and Vernonia adoensis chondrillasterol purified from acetone extract [117].
Furthermore, the hypermucoviscosity phenotype of Kpn strains plays an essential role in antibiotic tolerance, as the extracellular matrix, composed of proteins, DNA, lipopolysaccharides, and lipids, acts as a protective barrier that prevents penetration by antibiotics, limiting the design of new therapies. Although many plant extracts have shown excellent results against Kpn, the poor knowledge of biofilms, hypermucoviscosity ecology, and physiology limits the application of these components.
Many antibacterial components cannot cross this barrier and act against the pathogen. Essential oils are secondary metabolites of a lipophilic nature with antibacterial, antiviral, and insecticidal properties, as plants produce them to fight pest invasions and predators [130].
In 2019, Vasconcelos et al. reported that the essential oil obtained from Origanum vulgare L. effectively inhibited KPC-Kpn strains Cinnamomum camphora essential oil is another plant-origin compound that is effective against MDR-Kpn strains [114]. In addition, essential oils from Thymus vulgaris and Syzygium aromaticum showed efficiency against KPC-Kpn strains individually and mixed with chitosan to form nanoemulsions, suggesting the use of these treatments to combat brain and central nervous system infections [131].
A combination of natural compounds has also been explored to determine their potential use to fight against Kpn disease. For example, the antibiotic effect of Melaleuca alternifolia against KPC-Kpn strains was studied alone as well as in combination with meropenem, amikacin, and colistin, showing that these compounds have a synergistic effect against KPC-Kpn strains [132]. Positive antibacterial results were also observed when using Cinnamomum burmanii, Mentha piperita, Thymus vulgaris, Camellia japonica, Artemisia herba-alba, and Thymus algeriensis essential oils [123,124,125][123][124][125]. Furthermore, combining M. alternifolia and T. vulgaris essential oils diminished biofilm formation [120]. Researchers' group found that Thymus vulgaris, Mentha piperita, Rosmarinus officinalis, and Curcuma longa essential oils effectively against Kpn clinical strains, affecting the hypermucoviscosity phenotype. These findings indicate that essential oils are the best choice to combat mucus production and biofilm-forming strains [121]. However, more studies on essential oils are needed to investigate their safety profile, characterise their mechanism of action, optimise their dose, and observe possible adverse effects in human cells.
In addition to essential oils, bioactive compounds obtained from essential oil extraction were found to have antimicrobial activity. For example, the phenolic monoterpene carvacrol, derived from Thymus spp., showed significant inhibitory effects against MDR-Kpn strains in a murine model [133]. Additionally, eugenol treatment affected biofilm formation [134]. The antibacterial effect of cinnamaldehyde and eugenol was demonstrated against KpC strains in vitro and in a murine model [135]. β-Sitosterol isolated from Kalanchoe tomentosa and elemicin isolated from Myristica fragans acted effectively against MDR-Kpn strains [136,137][136][137].
POCs possess a wide range of mechanisms of action against bacteria, such as breakdown of the membrane of the cell, inhibition of wall synthesis, synthesis inhibition and denaturation of proteins, an increase in reactive oxygen species (ROS), alterations to efflux pumps, and the use of metal chelators. Although most POCs’ mechanisms of action against Kpn are insufficiently understood, the mechanisms are well characterised for some POCs. The essential oil of Juniperus rigida affects cell permeability and damages the integrity of the membranes of cells, as demonstrated by the leaking of proteins, RNA, and DNA, as well as morphological changes observed using scanning electron microscopy [126]. Plectranthus amboinicus essential oil alters membrane integrity and inhibits capsule expression [127]. Lavandula angustifolia essential oil and Cinnamomum verum oil disrupt bacterial membranes by generating oxidative stress. These oils oxidise the outer membrane, enabling the influx of generated ROS into the cells, causing damage to the cells and eventually death [128,138][128][138]. Methanol extract of Syzygium cumini has an inhibitory effect on quorum-sensing-regulated violacein production, biofilm production, and exopolysaccharides synthesis [139]. Hexane extract of Halimeda discoidea induces different morphological changes during different treatment periods, suggesting that the bioactive compound of the extract has different target sites and killing mechanisms, and their mixture makes the extract effective against multidrug-resistant bacteria [140].
5. Treatment Development
A severe health problem in many countries is the administration of antibiotics without susceptibility tests, resulting in high rates of antibiotic-resistant strains [97,98][97][98]. Now, in a “post-antibiotic era”, new treatment options are imperative. Immunotherapy is one of the most promising strategies for treating antibiotic-resistant pathogens, including MDR-Kpn strains. Passive immunisation using Kpn proteins has effective results. However, the high variability of CPS proteins is a limitation of this type of treatment [99]. Another treatment strategy is to use bacteriophages as Trojan horses that cause bacterial lysis. The Caudoviricetes, Myoviridae, Siphoviridae, Podoviridae, and Ackermannviridae families have been identified to infect the Kpn strain [100]. Mechanistically, bacteriophages create pores in the cell membrane and degrade peptidoglycan, disrupting cell membrane integrity [101]. Intraperitoneally administered in a murine model, bacteriophages showed promising results for therapeutic and prophylactic use against Kpn disease [102,103][102][103]. The limitations of this therapy include microbiome alteration and high production costs, and the safety implications in humans are still under investigation. Nevertheless, preliminary findings have shown effective results from the use of combined therapies with antibiotics and bacteriophages against Kpn strains [104]. A priority in the effort against MDR bacteria is developing new efficient therapies or complementing existing treatments. Plant-origin components (POCs) have received particular attention because of their properties, economic viability, and efficacy against bacterial disease. POCs provide an alternative as antibiotic adjuvants, enhancing antibiotic activity.6. The Potential of Plant-Origin Components as a New Treatment Source
Medicinal plants are still used in many societies. The continued use of plants to combat diseases in these societies is based on tradition, economic limitations, and religion. Even though modern medicine has had enormous advances, these sources have been retaken to identify new treatments in the fight against multiple-antibiotic-resistant Kpn diseases. Since 35,000 to 70,000 plant species possess therapeutic properties, their use to treat various conditions is a promising approach [105]. Plants produce secondary metabolites in response to environmental factors such as predators, abiotic stress, and interspecific interactions; many of these metabolites could be helpful for pharmaceutic development [106]. Although many POCs and molecules have been isolated, information regarding mechanisms of action, signal transduction, and potential to combat specific diseases is poorly understood. Using different methodological approaches, POCs can be extracted from many parts of plants, such as leaves, seeds, fruits, flowers, roots, wood, etc. (Figure 5).
Figure 5.
Plant-origin compound extraction methods and application.
Table 2.
Plant-origin compounds that present antibacterial activity against
Klebsiella pneumoniae
strains.
| Plant | Type of Compound | Activity | Reference | ||||
|---|---|---|---|---|---|---|---|
| Momordica charantia | Ethanolic leaf extract Ethyl acetate leaf extract |
MIC = 625 µg/mL MIC = 156.2 µg/mL |
[108] | ||||
| Skimmia anquetilia | Ethyl acetate root extract | Zone diameter = 17.0 ± 1.0 mm MIC = 8 mg/mL |
[109,110][109][110] | ||||
| Bacopa monnieri | Ethanolic leaf extract Methanolic leaf extracts |
Zone diameter =23.0 ± 0.4 mm Zone diameter = 25.0 ± 0.5 mm |
[109,110][109][110] | ||||
| Paeonia officinalis | |||||||
| Thymus vulgaris | |||||||
| In vivo using albino Holtzman rats | |||||||
| While the 28-day oral toxicity test indicated that the no-observed-adverse-effect level (NOAEL) was more than 250 mg/kg/day, | Thymus vulgaris | had moderate oral toxicity. | [158] | ||||
| Vernonia adoensis | Chondrillasterol purified from acetone extract | Reduces bacterial growth by 38% at 100 µg/mL | [ | ||||
| Syzygium aromaticum | In vitro against human normal dermal fibroblasts | Syzygium aromaticum oil cytotoxicity is dose-dependent at a concentration of 0.03%. | 117] | ||||
| [ | 159 | ] | Origanum vulgare | ||||
| Cinnamomum verum | Essential oil | Zone diameter = 21 mm | In vivo using G. mellonella larvae MIC = 0.059% (v/v) |
[ | C. verum leaf EO was non-toxic in the experimental model118] | ||
| [ | 160 | ] | Cinnamomum camphora | Essential oil | MIC = 6.25% (v/v) MBC = 12.5% (v/v | ||
| Mentha piperita | ) | In vivo and in vitro assays | Several (but not all) short-term and subchronic oral studies noted cystlike lesions in the cerebellum in rats that were given doses of Mentha piperita[119] | ||||
| oil containing pulegone, pulegone alone, or large amounts (>200 mg/kg/day) of menthone. Thus, it is safe if the concentration of pulegone in these ingredients does not exceed 1%. | [ | 161 | ] | Thymus vulgaris | Essential oil | ||
| Rosmarinus officinalis | Zone diameter of 21–30 mm | In vivo in Swiss albino miceMIC vary from 1 to 16 µg/mL | [120] | ||||
| No significative changes were reported in relative liver, spleen, heart, or lung size and morphology, and there were changes in clinical chemistry parameters | [ | 162 | ] | ||||
| Curcuma longa | MIC = 0.15% ( | v/v) |
In vivo in Wistar albino ratsMBC = 0.45% (v/v) | [121] | |||
| No clinical signs of toxicity were observed in any of the treated or control mice at a dose of 5000 mg/kg body weight | [ | 163 | ]v | Syzygium aromaticum | Essential oil | MIC = 0.078% (v/v) MBC = 0.156% (v/v) |
[122,123,124,125][122][123][124][125] |
| Melaleuca alternifolia | Essential oil | Zone diameter of 31–40 mm MIC varies from 0.5 to 4.0 µg/mL |
[120] | ||||
| Cinnamomum burmanii | Essential oil | MIC = 0.078% (v/v) MBC = 0.156% (v/v) |
[122,123,124,125][122][123][124][125] | ||||
| Cinnamomum verum | Essential oil | MIC = 0.5 mg/mL MBC = 1.0 mg/mL |
[122,123,124,125][122][123][124][125] | ||||
| Mentha piperita | Essential oil | Zone diameter of 21–30 mm MIC varies from 8 to 128 µg/mL |
[120] | ||||
| MIC = 0.60% (v/v) MBC = 1.25% (v/v) |
[121] | ||||||
| Camellia japonica | Essential oil | Zone diameter of 16 mm at 60 µg/mL MIC and MBC = 50 µg/mL |
[122] | ||||
| Rosmarinus officinalis | Essential oil | MIC = 0.45% (v/v) MBC = 3.75% (v/v) |
[121] | ||||
| Curcuma longa | Essential oil | MIC = 2.55% (v/v) MBC = 6.265% (v/v) |
[121] | ||||
| Juniperus rigida | Essential oil | Zone diameter = 16 ± 0.25 mm MIC and MBC = 3.125 mg/mL |
[126] | ||||
| Plectranthus amboinicus | Essential oil | MIC and MBC = 0.08% (700 µg/mL) | [127] | ||||
| Lavandula angustifolia | Essential oil | MIC = 10% | [128] |
References
- Friedlaender, C. Ueber Die Schizomyceten Bei Der Acuten Fibrösen Pneumonie. Arch. Pathol. Anat. Physiol. Klin. Med. 1882, 87, 319–324.
- Brisse, S.; Grimont, F.; Grimont, P.A.D. The Genus Klebsiella. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 159–196.
- Long, S.W.; Linson, S.E.; Ojeda Saavedra, M.; Cantu, C.; Davis, J.J.; Brettin, T.; Olsen, R.J. Whole-Genome Sequencing of Human Clinical Klebsiella pneumoniae Isolates Reveals Misidentification and Misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2017, 2, 10–128.
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population Genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359.
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic Analysis of Diversity, Population Structure, Virulence, and Antimicrobial Resistance in Klebsiella pneumoniae, an Urgent Threat to Public Health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581.
- Catalán-Nájera, J.C.; Garza-Ramos, U.; Barrios-Camacho, H. Hypervirulence and Hypermucoviscosity: Two Different but Complementary Klebsiella spp. Phenotypes? Virulence 2017, 8, 1111–1123.
- Chen, L.; Kreiswirth, B.N. Convergence of Carbapenem-Resistance and Hypervirulence in Klebsiella pneumoniae. Lancet Infect. Dis. 2018, 18, 2–3.
- Tang, M.; Kong, X.; Hao, J.; Liu, J. Epidemiological Characteristics and Formation Mechanisms of Multidrug-Resistant Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2020, 11, 581543.
- Choby, J.E.; Howard-Anderson, J.; Weiss, D.S. Hypervirulent Klebsiella pneumoniae—Clinical and Molecular Perspectives. J. Intern. Med. 2020, 287, 283–300.
- Hennequin, C.; Robin, F. Correlation between Antimicrobial Resistance and Virulence in Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 333–341.
- Stojowska-Swędrzyńska, K.; Łupkowska, A.; Kuczyńska-Wiśnik, D.; Laskowska, E. Antibiotic Heteroresistance in Klebsiella pneumoniae. Int. J. Mol. Sci. 2021, 23, 449.
- Savithramma, N.; Linga Rao, M.; Suhrulatha, D. Screening of Selected Medicinal Plants for Secondary Metabolites. Middle East J. Sci. Res. 2011, 8, 579–584.
- Struve, C.; Bojer, M.; Krogfelt, K.A. Characterization of Klebsiella pneumoniae Type 1 Fimbriae by Detection of Phase Variation during Colonization and Infection and Impact on Virulence. Infect. Immun. 2008, 76, 4055–4065.
- Alcántar-Curiel, M.D.; Blackburn, D.; Saldaña, Z.; Gayosso-Vázquez, C.; Iovine, N.M.; De la Cruz, M.A.; Girón, J.A. Multi-Functional Analysis of Klebsiella pneumoniae Fimbrial Types in Adherence and Biofilm Formation. Virulence 2013, 4, 129–138.
- Struve, C.; Bojer, M.; Krogfelt, K.A. Identification of a Conserved Chromosomal Region Encoding Klebsiella pneumoniae Type 1 and Type 3 Fimbriae and Assessment of the Role of Fimbriae in Pathogenicity. Infect. Immun. 2009, 77, 5016–5024.
- Opoku-Temeng, C.; Kobayashi, S.D.; DeLeo, F.R. Klebsiella pneumoniae Capsule Polysaccharide as a Target for Therapeutics and Vaccines. Comput. Struct. Biotechnol. J. 2019, 17, 1360–1366.
- Hsu, C.-R.; Pan, Y.-J.; Liu, J.-Y.; Chen, C.-T.; Lin, T.-L.; Wang, J.-T. Klebsiella pneumoniae Translocates across the Intestinal Epithelium via Rho GTPase- and Phosphatidylinositol 3-Kinase/Akt-Dependent Cell Invasion. Infect. Immun. 2015, 83, 769–779.
- DeLeo, F.R.; Chen, L.; Porcella, S.F.; Martens, C.A.; Kobayashi, S.D.; Porter, A.R.; Chavda, K.D.; Jacobs, M.R.; Mathema, B.; Olsen, R.J.; et al. Molecular Dissection of the Evolution of Carbapenem-Resistant Multilocus Sequence Type 258 Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 2014, 111, 4988–4993.
- Regueiro, V.; Campos, M.A.; Pons, J.; Albertí, S.; Bengoechea, J.A. The Uptake of a Klebsiella pneumoniae Capsule Polysaccharide Mutant Triggers an Inflammatory Response by Human Airway Epithelial Cells. Microbiology 2006, 152, 555–566.
- Pan, Y.-J.; Lin, T.-L.; Hsu, C.-R.; Wang, J.-T. Use of a Dictyostelium Model for Isolation of Genetic Loci Associated with Phagocytosis and Virulence in Klebsiella pneumoniae. Infect. Immun. 2011, 79, 997–1006.
- You, H.S.; Lee, S.H.; Kang, S.S.; Hyun, S.H. OmpA of Klebsiella pneumoniae ATCC 13883 Induces Pyroptosis in HEp-2 Cells, Leading to Cell-Cycle Arrest and Apoptosis. Microbes Infect. 2020, 22, 432–440.
- Rocker, A.; Lacey, J.A.; Belousoff, M.J.; Wilksch, J.J.; Strugnell, R.A.; Davies, M.R.; Lithgow, T. Global Trends in Proteome Remodeling of the Outer Membrane Modulate Antimicrobial Permeability in Klebsiella pneumoniae. mBio 2020, 11, 10–128.
- Merino, S.; Camprubi, S.; Alberti, S.; Benedi, V.J.; Tomas, J.M. Mechanisms of Klebsiella pneumoniae Resistance to Complement-Mediated Killing. Infect. Immun. 1992, 60, 2529–2535.
- Lawlor, M.S.; O’Connor, C.; Miller, V.L. Yersiniabactin Is a Virulence Factor for Klebsiella pneumoniae during Pulmonary Infection. Infect. Immun. 2007, 75, 1463–1472.
- Bachman, M.A.; Oyler, J.E.; Burns, S.H.; Caza, M.; Lépine, F.; Dozois, C.M.; Weiser, J.N. Klebsiella pneumoniae Yersiniabactin Promotes Respiratory Tract Infection through Evasion of Lipocalin 2. Infect. Immun. 2011, 79, 3309–3316.
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451.
- Bachman, M.A.; Lenio, S.; Schmidt, L.; Oyler, J.E.; Weiser, J.N. Interaction of Lipocalin 2, Transferrin, and Siderophores Determines the Replicative Niche of Klebsiella pneumoniae during Pneumonia. mBio 2012, 3, 10–128.
- Holden, V.I.; Wright, M.S.; Houle, S.; Collingwood, A.; Dozois, C.M.; Adams, M.D.; Bachman, M.A. Iron Acquisition and Siderophore Release by Carbapenem-Resistant Sequence Type 258 Klebsiella pneumoniae. mSphere 2018, 3, e00125-18.
- Chen, Y.-T.; Chang, H.-Y.; Lai, Y.-C.; Pan, C.-C.; Tsai, S.-F.; Peng, H.-L. Sequencing and Analysis of the Large Virulence Plasmid PLVPK of Klebsiella pneumoniae CG43. Gene 2004, 337, 189–198.
- Russo, T.A.; Olson, R.; MacDonald, U.; Metzger, D.; Maltese, L.M.; Drake, E.J.; Gulick, A.M. Aerobactin Mediates Virulence and Accounts for Increased Siderophore Production under Iron-Limiting Conditions by Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 2014, 82, 2356–2367.
- Sarris, P.F.; Zoumadakis, C.; Panopoulos, N.J.; Scoulica, E.V. Distribution of the Putative Type VI Secretion System Core Genes in Klebsiella spp. Infect. Genet. Evol. 2011, 11, 157–166.
- Liu, L.; Ye, M.; Li, X.; Li, J.; Deng, Z.; Yao, Y.-F.; Ou, H.Y. Identification and Characterization of an Antibacterial Type VI Secretion System in the Carbapenem-Resistant Strain Klebsiella pneumoniae HS11286. Front. Cell Infect. Microbiol. 2017, 7, 442.
- Liu, W.; Li, M.; Cao, S.; Ishaq, H.M.; Zhao, H.; Yang, F.; Liu, L. The Biological and Regulatory Role of Type VI Secretion System of Klebsiella pneumoniae. Infect. Drug Resist. 2023, 16, 6911–6922.
- Lu, M.-C.; Chen, Y.-T.; Chiang, M.-K.; Wang, Y.-C.; Hsiao, P.-Y.; Huang, Y.-J.; Lin, C.-T.; Cheng, C.-C.; Liang, C.-L.; Lai, Y.-C. Colibactin Contributes to the Hypervirulence of Pks(+) K1 CC23 Klebsiella pneumoniae in Mouse Meningitis Infections. Front. Cell. Infect. Microbiol. 2017, 7, 103.
- Podschun, R.; Ullmann, U. Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors. Clin. Microbiol. Rev. 1998, 11, 589–603.
- Seguel, M.; Gottdenker, N.L.; Colegrove, K.; Johnson, S.; Struve, C.; Howerth, E.W. Hypervirulent Klebsiella pneumoniae in California Sea Lions (Zalophus californianus): Pathologic Findings in Natural Infections. Vet. Pathol. 2017, 54, 846–850.
- Du, Q.; Pan, F.; Wang, C.; Yu, F.; Shi, Y.; Liu, W.; Li, Z.; He, P.; Han, D.; Zhang, H. Nosocomial Dissemination of Hypervirulent Klebsiella pneumoniae with High-Risk Clones among Children in Shanghai. Front. Cell. Infect. Microbiol. 2022, 12, 984180.
- Siu, L.K.; Chang, F.-Y. Klebsiella pneumoniae Liver Abscesses--Authors’ Reply. Lancet Infect. Dis. 2013, 13, 393.
- Zhao, J.; Huo, T.; Luo, X.; Lu, F.; Hui, S.; Yang, B. Klebsiella pneumoniae-Related Brain Abscess and Meningitis in Adults: Case Report. Medicine 2022, 101, e28415.
- Zandi, R.; Talebi, S.; Sheibani, S.; Ehsani, A. Klebsiella pneumoniae and Enterobacter cloacae Induced Septic Arthritis in a Healthy Adolescent: A Rare Case Report. Hip Pelvis 2022, 34, 185–190.
- Kritikos, A.; Manuel, O. Bloodstream Infections after Solid-Organ Transplantation. Virulence 2016, 7, 329–340.
- Bodro, M.; Sabé, N.; Tubau, F.; Lladó, L.; Baliellas, C.; Roca, J.; Cruzado, J.M.; Carratalà, J. Risk Factors and Outcomes of Bacteremia Caused by Drug-Resistant ESKAPE Pathogens in Solid-Organ Transplant Recipients. Transplantation 2013, 96, 843–849.
- Zhang, Y.; Guo, L.-Y.; Song, W.-Q.; Wang, Y.; Dong, F.; Liu, G. Risk Factors for Carbapenem-Resistant K. pneumoniae Bloodstream Infection and Predictors of Mortality in Chinese Paediatric Patients. BMC Infect. Dis. 2018, 18, 248.
- Caballero, S.; Carter, R.; Ke, X.; Sušac, B.; Leiner, I.M.; Kim, G.J.; Miller, L.; Ling, L.; Manova, K.; Pamer, E.G. Distinct but Spatially Overlapping Intestinal Niches for Vancomycin-Resistant Enterococcus faecium and Carbapenem-Resistant Klebsiella pneumoniae. PLoS Pathog. 2015, 11, e1005132.
- Abril, D.; Vergara, E.; Palacios, D.; Leal, A.L.; Marquez-Ortiz, R.A.; Madroñero, J.; Corredor Rozo, Z.L.; De La Rosa, Z.; Nieto, C.A.; Vanegas, N.; et al. Within Patient Genetic Diversity of BlaKPC Harboring Klebsiella pneumoniae in a Colombian Hospital and Identification of a New NTEKPC Platform. Sci. Rep. 2021, 11, 21409.
- Gaspari, R.; Spinazzola, G.; Teofili, L.; Avolio, A.W.; Fiori, B.; Maresca, G.M.; Spanu, T.; Nicolotti, N.; de Pascale, G.; Antonelli, M. Protective Effect of SARS-CoV-2 Preventive Measures against ESKAPE and Escherichia coli Infections. Eur. J. Clin. Investig. 2021, 51, e13687.
- Micozzi, A.; Assanto, G.M.; Cesini, L.; Minotti, C.; Cartoni, C.; Capria, S.; Ciotti, G.; Alunni Fegatelli, D.; Donzelli, L.; Martelli, M.; et al. Reduced Transmission of Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae (KPC-KP) in Patients with Haematological Malignancies Hospitalized in an Italian Hospital during the COVID-19 Pandemic. JAC Antimicrob. Resist. 2021, 3, dlab167.
- Cerulli Irelli, E.; Morano, A.; Di Bonaventura, C. Reduction in Nosocomial Infections during the COVID-19 Era: A Lesson to Be Learned. Updates Surg. 2021, 73, 785–786.
- Rawson, T.M.; Zhu, N.; Ranganathan, N.; Gilchrist, M.; Satta, G.; Cooke, G.; Holmes, A. Bacterial and Fungal Co-Infection in Individuals with Coronavirus: A Rapid Review to Support COVID-19 Antimicrobial Prescribing Timothy. Clin. Infect. Dis. 2020, 71, 2459–2468.
- Salazar-Vizcaya, L.; Atkinson, A.; Kronenberg, A.; Plüss-Suard, C.; Kouyos, R.D.; Kachalov, V.; Troillet, N.; Marschall, J.; Sommerstein, R. The Impact of Public Health Interventions on the Future Prevalence of ESBL-Producing Klebsiella pneumoniae: A Population Based Mathematical Modelling Study. BMC Infect. Dis. 2022, 22, 487.
- Cejas, D.; Magariños, F.; Elena, A.; Ferrara, M.; Ormazábal, C.; Yernazian, M.V.; Gutkind, G.; Radice, M. Emergence and Clonal Expansion of Klebsiella pneumoniae ST307, Simultaneously Producing KPC-3 and NDM-1. Rev. Argent Microbiol. 2022, 54, 288–292.
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Emerging Antimicrobial-Resistant High-Risk Klebsiella pneumoniae Clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20.
- Hernández-García, M.; Castillo-Polo, J.A.; Cordero, D.G.; Pérez-Viso, B.; García-Castillo, M.; Saez de la Fuente, J.; Morosini, M.I.; Cantón, R.; Ruiz-Garbajosa, P. Impact of Ceftazidime-Avibactam Treatment in the Emergence of Novel KPC Variants in the ST307-Klebsiella pneumoniae High-Risk Clone and Consequences for Their Routine Detection. J. Clin. Microbiol. 2022, 60, e0224521.
- Mataseje, L.F.; Chen, L.; Peirano, G.; Fakharuddin, K.; Kreiswith, B.; Mulvey, M.; Pitout, J.D.D. Klebsiella pneumoniae ST147: And Then There Were Three Carbapenemases. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1467–1472.
- McKenna, M. Antibiotic Resistance: The Last Resort. Nature 2013, 499, 394–396.
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and Resistance Mechanisms of Antibiotics: A Guide for Clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300.
- Zhang, F.; Cheng, W. The Mechanism of Bacterial Resistance and Potential Bacteriostatic Strategies. Antibiotics 2022, 11, 1215.
- Srinivasan, V.B.; Venkataramaiah, M.; Mondal, A.; Vaidyanathan, V.; Govil, T.; Rajamohan, G. Functional Characterization of a Novel Outer Membrane Porin KpnO, Regulated by PhoBR Two-Component System in Klebsiella pneumoniae NTUH-K2044. PLoS ONE 2012, 7, e41505.
- Srinivasan, V.B.; Rajamohan, G. KpnEF, a New Member of the Klebsiella pneumoniae Cell Envelope Stress Response Regulon, Is an SMR-Type Efflux Pump Involved in Broad-Spectrum Antimicrobial Resistance. Antimicrob. Agents Chemother. 2013, 57, 4449–4462.
- Machulska, M.; Baraniak, A.; Żak, I.; Bojarska, K.; Żabicka, D.; Sowa-Sierant, I.; Hryniewicz, W.; Gniadkowski, M. KPC-2-Producing Klebsiella pneumoniae ST11 in a Children’s Hospital in Poland. Pol. J. Microbiol. 2017, 66, 401–404.
- Kliebe, C.; Nies, B.A.; Meyer, J.F.; Tolxdorff-Neutzling, R.M.; Wiedemann, B. Evolution of Plasmid-Coded Resistance to Broad-Spectrum Cephalosporins. Antimicrob. Agents Chemother. 1985, 28, 302–307.
- Sirot, D.; Sirot, J.; Labia, R.; Morand, A.; Courvalin, P.; Darfeuille-Michaud, A.; Perroux, R.; Cluzel, R. Transferable Resistance to Third-Generation Cephalosporins in Clinical Isolates of Klebsiella pneumoniae: Identification of CTX-1, a Novel β-Lactamase. J. Antimicrob. Chemother. 1987, 20, 323–334.
- Calbo, E.; Garau, J. The Changing Epidemiology of Hospital Outbreaks Due to ESBL-Producing Klebsiella pneumoniae: The CTX-M-15 Type Consolidation. Future Microbiol. 2015, 10, 1063–1075.
- Evans, B.A.; Amyes, S.G.B. OXA β-Lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263.
- Bradford, P.A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951.
- Zamorano, L.; Miró, E.; Juan, C.; Gómez, L.; Bou, G.; González-López, J.J.; Martínez-Martínez, L.; Aracil, B.; Conejo, M.C.; Oliver, A.; et al. Mobile Genetic Elements Related to the Diffusion of Plasmid-Mediated AmpC β-Lactamases or Carbapenemases from Enterobacteriaceae: Findings from a Multicenter Study in Spain. Antimicrob. Agents Chemother. 2015, 59, 5260–5266.
- Haruta, S.; Yamaguchi, H.; Yamamoto, E.T.; Eriguchi, Y.; Nukaga, M.; O’Hara, K.; Sawai, T. Functional Analysis of the Active Site of a Metallo-β-Lactamase Proliferating in Japan. Antimicrob. Agents Chemother. 2000, 44, 2304–2309.
- Lee, C.-R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895.
- Molton, J.S.; Tambyah, P.A.; Ang, B.S.P.; Ling, M.L.; Fisher, D.A. The Global Spread of Healthcare-Associated Multidrug-Resistant Bacteria: A Perspective From Asia. Clin. Infect. Dis. 2013, 56, 1310–1318.
- Leavitt, A.; Chmelnitsky, I.; Colodner, R.; Ofek, I.; Carmeli, Y.; Navon-Venezia, S. Ertapenem Resistance among Extended-Spectrum-β-Lactamase-Producing Klebsiella pneumoniae Isolates. J. Clin. Microbiol. 2009, 47, 969–974.
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.A.; Albertí, S. Klebsiella pneumoniae AcrAB Efflux Pump Contributes to Antimicrobial Resistance and Virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183.
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029.
- Doi, Y.; Wachino, J.I.; Arakawa, Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. 2016, 30, 523–537.
- Deguchi, T.; Fukuoka, A.; Yasuda, M.; Nakano, M.; Ozeki, S.; Kanematsu, E.; Nishino, Y.; Ishihara, S.; Ban, Y.; Kawada, Y. Alterations in the GyrA Subunit of DNA Gyrase and the ParC Subunit of Topoisomerase IV in Quinolone-Resistant Clinical Isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 1997, 41, 699–701.
- Guillard, T.; de Jong, A.; Limelette, A.; Lebreil, A.L.; Madoux, J.; de Champs, C. Characterization of Quinolone Resistance Mechanisms in Enterobacteriaceae Recovered from Diseased Companion Animals in Europe. Vet. Microbiol. 2016, 194, 23–29.
- Nam, Y.S.; Cho, S.Y.; Yang, H.Y.; Park, K.S.; Jang, J.-H.; Kim, Y.-T.; Jeong, J.; Suh, J.-T.; Lee, H.J. Investigation of Mutation Distribution in DNA Gyrase and Topoisomerase IV Genes in Ciprofloxacin-Non-Susceptible Enterobacteriaceae Isolated from Blood Cultures in a Tertiary Care University Hospital in South Korea, 2005–2010. Int. J. Antimicrob. Agents 2013, 41, 126–129.
- Wong, M.H.Y.; Chan, E.W.C.; Chen, S. Evolution and Dissemination of OqxAB-Like Efflux Pumps, an Emerging Quinolone Resistance Determinant among Members of Enterobacteriaceae. Antimicrob. Agents Chemother. 2015, 59, 3290–3297.
- Jacoby, G.A.; Strahilevitz, J.; Hooper, D.C. Plasmid-Mediated Quinolone Resistance. Microbiol. Spectr. 2014, 2, 475–503.
- Ruiz, E.; Sáenz, Y.; Zarazaga, M.; Rocha-Gracia, R.; Martínez-Martínez, L.; Arlet, G.; Torres, C. Qnr, Aac(6′)-Ib-Cr and QepA Genes in Escherichia Coli and Klebsiella spp.: Genetic Environments and Plasmid and Chromosomal Location. J. Antimicrob. Chemother. 2012, 67, 886–897.
- De Majumdar, S.; Yu, J.; Fookes, M.; McAteer, S.P.; Llobet, E.; Finn, S.; Spence, S.; Monaghan, A.; Kissenpfennig, A.; Ingram, R.J.; et al. Elucidation of the RamA Regulon in Klebsiella pneumoniae Reveals a Role in LPS Regulation. PLoS Pathog. 2015, 11, e1004627.
- Clements, A.; Tull, D.; Jenney, A.W.; Farn, J.L.; Kim, S.-H.; Bishop, R.E.; McPhee, J.B.; Hancock, R.E.W.; Hartland, E.L.; Pearse, M.J.; et al. Secondary Acylation of Klebsiella pneumoniae Lipopolysaccharide Contributes to Sensitivity to Antibacterial Peptides. J. Biol. Chem. 2007, 282, 15569–15577.
- Mitrophanov, A.Y.; Jewett, M.W.; Hadley, T.J.; Groisman, E.A. Evolution and Dynamics of Regulatory Architectures Controlling Polymyxin B Resistance in Enteric Bacteria. PLoS Genet. 2008, 4, e1000233.
- Llobet, E.; Campos, M.A.; Giménez, P.; Moranta, D.; Bengoechea, J.A. Analysis of the Networks Controlling the Antimicrobial-Peptide-Dependent Induction of Klebsiella pneumoniae Virulence Factors. Infect. Immun. 2011, 79, 3718–3732.
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168.
- Jayol, A.; Poirel, L.; Brink, A.; Villegas, M.-V.; Yilmaz, M.; Nordmann, P. Resistance to Colistin Associated with a Single Amino Acid Change in Protein PmrB among Klebsiella pneumoniae Isolates of Worldwide Origin. Antimicrob. Agents Chemother. 2014, 58, 4762–4766.
- De Majumdar, S.; Veleba, M.; Finn, S.; Fanning, S.; Schneiders, T. Elucidating the Regulon of Multidrug Resistance Regulator RarA in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2013, 57, 1603–1609.
- Osei Sekyere, J.; Govinden, U.; Bester, L.A.; Essack, S.Y. Colistin and Tigecycline Resistance in Carbapenemase-Producing Gram-Negative Bacteria: Emerging Resistance Mechanisms and Detection Methods. J. Appl. Microbiol. 2016, 121, 601–617.
- Villa, L.; Feudi, C.; Fortini, D.; García-Fernández, A.; Carattoli, A. Genomics of KPC-Producing Klebsiella pneumoniae Sequence Type 512 Clone Highlights the Role of RamR and Ribosomal S10 Protein Mutations in Conferring Tigecycline Resistance. Antimicrob. Agents Chemother. 2014, 58, 1707–1712.
- Nielsen, L.E.; Snesrud, E.C.; Onmus-Leone, F.; Kwak, Y.I.; Avilés, R.; Steele, E.D.; Sutter, D.E.; Waterman, P.E.; Lesho, E.P. IS5 Element Integration, a Novel Mechanism for Rapid in Vivo Emergence of Tigecycline Nonsusceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2014, 58, 6151–6156.
- Wilson, D.N.; Hauryliuk, V.; Atkinson, G.C.; O’Neill, A.J. Target Protection as a Key Antibiotic Resistance Mechanism. Nat. Rev. Microbiol. 2020, 18, 637–648.
- Tran, J.H.; Jacoby, G.A.; Hooper, D.C. Interaction of the Plasmid-Encoded Quinolone Resistance Protein Qnr with Escherichia coli DNA Gyrase. Antimicrob. Agents Chemother. 2005, 49, 118–125.
- Amereh, F.; Arabestani, M.R.; Hosseini, S.M.; Shokoohizadeh, L. Association of Qnr Genes and OqxAB Efflux Pump in Fluoroquinolone-Resistant Klebsiella pneumoniae Strains. Int. J. Microbiol. 2023, 2023, 9199108.
- Gunn, J.S. Bacterial Modification of LPS and Resistance to Antimicrobial Peptides. J. Endotoxin Res. 2001, 7, 57–62.
- Mazzariol, A.; Zuliani, J.; Cornaglia, G.; Rossolini, G.M.; Fontana, R. AcrAB Efflux System: Expression and Contribution to Fluoroquinolone Resistance in Klebsiella spp. Antimicrob. Agents Chemother. 2002, 46, 3984–3986.
- Ping, Y.; Ogawa, W.; Kuroda, T.; Tsuchiya, T. Gene Cloning and Characterization of KdeA, a Multidrug Efflux Pump from Klebsiella pneumoniae. Biol. Pharm. Bull. 2007, 30, 1962–1964.
- Vuotto, C.; Longo, F.; Balice, M.P.; Donelli, G.; Varaldo, P.E. Antibiotic Resistance Related to Biofilm Formation in Klebsiella pneumoniae. Pathogens 2014, 3, 743–758.
- Hadi, U.; Kuntaman, K.; Qiptiyah, M.; Paraton, H. Problem of Antibiotic Use and Antimicrobial Resistance in Indonesia: Are We Really Making Progress? Indones. J. Trop. Infect. Dis. 2013, 4, 5.
- van der Meer, J.W.M.; Gyssens, I.C. Quality of Antimicrobial Drug Prescription in Hospital. Clin. Microbiol. Infect. 2001, 7, 12–15.
- Diago-Navarro, E.; Calatayud-Baselg, I.; Sun, D.; Khairallah, C.; Mann, I.; Ulacia-Hernando, A.; Sheridan, B.; Shi, M.; Fries, B.C. Antibody-Based Immunotherapy to Treat and Prevent Infection with Hypervirulent Klebsiella pneumoniae. Clin. Vaccine Immunol. 2017, 24, e00456-16.
- Herridge, W.P.; Shibu, P.; O’Shea, J.; Brook, T.C.; Hoyles, L. Bacteriophages of Klebsiella spp., Their Diversity and Potential Therapeutic Uses. J. Med. Microbiol. 2020, 69, 176–194.
- Domingo-Calap, P.; Delgado-Martínez, J. Bacteriophages: Protagonists of a Post-Antibiotic Era. Antibiotics 2018, 7, 66.
- Soleimani Sasani, M.; Eftekhar, F. Potential of a Bacteriophage Isolated from Wastewater in Treatment of Lobar Pneumonia Infection Induced by Klebsiella pneumoniae in Mice. Curr. Microbiol. 2020, 77, 2650–2655.
- Arena, F.; Menchinelli, G.; di Pilato, V.; Torelli, R.; Antonelli, A.; Henrici De Angelis, L.; Coppi, M.; Sanguinetti, M.; Rossolini, G.M. Resistance and Virulence Features of Hypermucoviscous Klebsiella pneumoniae from Bloodstream Infections: Results of a Nationwide Italian Surveillance Study. Front. Microbiol. 2022, 13, 983294.
- Verma, V.; Harjai, K.; Chhibber, S. Restricting Ciprofloxacin-Induced Resistant Variant Formation in Biofilm of Klebsiella pneumoniae B5055 by Complementary Bacteriophage Treatment. J. Antimicrob. Chemother. 2009, 64, 1212–1218.
- Veeresham, C. Natural Products Derived from Plants as a Source of Drugs. J. Adv. Pharm. Technol. Res. 2012, 3, 200.
- Yeshi, K.; Crayn, D.; Ritmejerytė, E.; Wangchuk, P. Plant Secondary Metabolites Produced in Response to Abiotic Stresses Has Potential Application in Pharmaceutical Product Development. Molecules 2022, 27, 313.
- Jha, A.K.; Sit, N. Extraction of Bioactive Compounds from Plant Materials Using Combination of Various Novel Methods: A Review. Trends Food Sci. Technol. 2022, 119, 579–591.
- Muribeca, A.d.J.B.; Gomes, P.W.P.; Paes, S.S.; da Costa, A.P.A.; Gomes, P.W.P.; de S. Viana, J.; Reis, J.D.E.; Pamplona, S.d.G.S.R.; Silva, C.; Bauermeister, A.; et al. Antibacterial Activity from Momordica charantia L. Leaves and Flavones Enriched Phase. Pharmaceutics 2022, 14, 1796.
- Nabi, M.; Tabassum, N.; Ganai, B.A. Phytochemical Screening and Antibacterial Activity of Skimmia anquetilia N.P. Taylor and Airy Shaw: A First Study from Kashmir Himalaya. Front. Plant Sci. 2022, 13, 937946.
- Mehta, J.; Utkarsh, K.; Fuloria, S.; Singh, T.; Sekar, M.; Salaria, D.; Rolta, R.; Begum, M.Y.; Gan, S.H.; Rani, N.N.I.M.; et al. Antibacterial Potential of Bacopa monnieri (L.) Wettst. and Its Bioactive Molecules against Uropathogens-An In Silico Study to Identify Potential Lead Molecule(s) for the Development of New Drugs to Treat Urinary Tract Infections. Molecules 2022, 27, 4971.
- Masota, N.E.; Ohlsen, K.; Schollmayer, C.; Meinel, L.; Holzgrabe, U. Isolation and Characterization of Galloylglucoses Effective against Multidrug-Resistant Strains of Escherichia coli and Klebsiella pneumoniae. Molecules 2022, 27, 5045.
- Elamary, R.B.; Albarakaty, F.M.; Salem, W.M. Efficacy of Acacia Nilotica Aqueous Extract in Treating Biofilm-Forming and Multidrug Resistant Uropathogens Isolated from Patients with UTI Syndrome. Sci. Rep. 2020, 10, 11125.
- Figueiredo, C.S.S.E.S.; Branco Santos, J.C.; Castro Junior, J.A.d.A.; Wakui, V.G.; Rodrigues, J.F.S.; Arruda, M.O.; Monteiro, A.d.S.; Monteiro-Neto, V.; Bomfim, M.R.Q.; Kato, L.; et al. Himatanthus drasticus Leaves: Chemical Characterization and Evaluation of Their Antimicrobial, Antibiofilm, Antiproliferative Activities. Molecules 2017, 22, 910.
- Thinina, A.; Karim, H.; Alia, M.; Karim, A. Evaluation and Quantification of the Inhibition of Biofilm and Planktonic Forms of Klebsiella pneumoniae by the Polyphenolic Extract of Pulicaria crispa. J. Adv. Pharm. Technol. Res. 2020, 11, 117.
- Sood, H.; Kumar, Y.; Gupta, V.K.; Arora, D.S. Bioprospecting the Antimicrobial, Antibiofilm and Antiproliferative Activity of Symplocos racemosa Roxb. Bark Phytoconstituents along with Their Biosafety Evaluation and Detection of Antimicrobial Components by GC-MS. BMC Pharmacol. Toxicol. 2020, 21, 78.
- Gato, E.; Rosalowska, A.; Martínez-Guitián, M.; Lores, M.; Bou, G.; Pérez, A. Anti-Adhesive Activity of a Vaccinium corymbosum Polyphenolic Extract Targeting Intestinal Colonization by Klebsiella pneumoniae. Biomed. Pharmacother. 2020, 132, 110885.
- Mozirandi, W.; Tagwireyi, D.; Mukanganyama, S. Evaluation of Antimicrobial Activity of Chondrillasterol Isolated from Vernonia adoensis (Asteraceae). BMC Complement. Altern. Med. 2019, 19, 249.
- Vasconcelos, N.G.; Croda, J.; Silva, K.E.; Motta, M.L.L.; Maciel, W.G.; Limiere, L.C.; Simionatto, S. Origanum vulgare L. Essential Oil Inhibits the Growth of Carbapenem-Resistant Gram-Negative Bacteria. Rev. Soc. Bras. Med. Trop. 2019, 52, e20180502.
- Mujawah, A.A.H.; Abdallah, E.M.; Alshoumar, S.A.; Alfarraj, M.I.; Alajel, S.M.I.; Alharbi, A.L.; Alsalman, S.A.; Alhumaydhi, F.A. GC-MS and in Vitro Antibacterial Potential of Cinnamomum camphora Essential Oil against Some Clinical Antibiotic-Resistant Bacterial Isolates. Eur. Rev. Med. Pharmacol. Sci. 2022, 26, 5372–5379.
- Iseppi, R.; di Cerbo, A.; Aloisi, P.; Manelli, M.; Pellesi, V.; Provenzano, C.; Camellini, S.; Messi, P.; Sabia, C. In Vitro Activity of Essential Oils Against Planktonic and Biofilm Cells of Extended-Spectrum β-Lactamase (ESBL)/Carbapenamase-Producing Gram-Negative Bacteria Involved in Human Nosocomial Infections. Antibiotics 2020, 9, 272.
- Romo-Castillo, M.; Flores-Bautista, V.A.; Guzmán-Gutiérrez, S.L.; Reyes-Chilpa, R.; León-Santiago, M.; Luna-Pineda, V.M. Synergy of Plant Essential Oils in Antibiotic Therapy to Combat Klebsiella pneumoniae Infections. Pharmaceuticals 2023, 16, 839.
- Ginting, E.V.; Retnaningrum, E.; Widiasih, D.A. Antibacterial Activity of Clove (Syzygium aromaticum) and Cinnamon (Cinnamomum burmannii) Essential Oil against Extended-Spectrum β-Lactamase-Producing Bacteria. Vet. World 2021, 14, 2206–2211.
- Wijesinghe, G.K.; Feiria, S.B.; Maia, F.C.; Oliveira, T.R.; Joia, F.; Barbosa, J.P.; Boni, G.C.; HÖfling, J.F. In-Vitro Antibacterial and Antibiofilm Activity of Cinnamomum verum Leaf Oil against Pseudomonas aeruginosa, Staphylococcus aureus and Klebsiella pneumoniae. Acad. Bras. Cienc. 2021, 93, e20201507.
- Ramachandran, G.; Rajivgandhi, G.N.; Murugan, S.; Alharbi, N.S.; Kadaikunnan, S.; Khaled, J.M.; Almanaa, T.N.; Manoharan, N.; Li, W.-J. Anti-Carbapenamase Activity of Camellia japonica Essential Oil against Isolated Carbapenem Resistant Klebsiella pneumoniae (MN396685). Saudi J. Biol. Sci. 2020, 27, 2269–2279.
- Vasconcelos, N.G.; Silva, K.E.; Croda, J.; Simionatto, S. Antibacterial Activity of Cinnamomum cassia L. Essential Oil in a Carbapenem- and Polymyxin-Resistant Klebsiella aerogenes Strain. Rev. Soc. Bras. Med. Trop. 2020, 53, e20200032.
- Meng, X.; Li, D.; Zhou, D.; Wang, D.; Liu, Q.; Fan, S. Chemical Composition, Antibacterial Activity and Related Mechanism of the Essential Oil from the Leaves of Juniperus rigida Sieb. et Zucc against Klebsiella pneumoniae. J. Ethnopharmacol. 2016, 194, 698–705.
- Gonçalves, T.B.; Braga, M.A.; de Oliveira, F.F.M.; Santiago, G.M.P.; Carvalho, C.B.M.; e Cabral, P.B.; Santiago, T.d.M.; Sousa, J.S.; Barros, E.B.; do Nascimento, R.F.; et al. Effect of Subinihibitory and Inhibitory Concentrations of Plectranthus amboinicus (Lour.) Spreng Essential Oil on Klebsiella pneumoniae. Phytomedicine 2012, 19, 962–968.
- Yang, S.-K.; Yusoff, K.; Thomas, W.; Akseer, R.; Alhosani, M.S.; Abushelaibi, A.; Lim, S.-H.-E.; Lai, K.-S. Lavender Essential Oil Induces Oxidative Stress Which Modifies the Bacterial Membrane Permeability of Carbapenemase Producing Klebsiella pneumoniae. Sci. Rep. 2020, 10, 819.
- Piperaki, E.-T.; Syrogiannopoulos, G.A.; Tzouvelekis, L.S.; Daikos, G.L. Klebsiella pneumoniae: Virulence, Biofilm and Antimicrobial Resistance. Pediatr. Infect. Dis. J. 2017, 36, 1002–1005.
- Chouhan, S.; Sharma, K.; Guleria, S. Antimicrobial Activity of Some Essential Oils—Present Status and Future Perspectives. Medicines 2017, 4, 58.
- Barrera-Ruiz, D.G.; Cuestas-Rosas, G.C.; Sánchez-Mariñez, R.I.; Álvarez-Ainza, M.L.; Moreno-Ibarra, G.M.; López-Meneses, A.K.; Plascencia-Jatomea, M.; Cortez-Rocha, M.O. Antibacterial Activity of Essential Oils Encapsulated in Chitosan Nanoparticles. Food Sci. Technol. 2020, 40, 568–573.
- Oliva, A.; D’Abramo, A.; D’Agostino, C.; Iannetta, M.; Mascellino, M.T.; Gallinelli, C.; Mastroianni, C.M.; Vullo, V. Synergistic Activity and Effectiveness of a Double-Carbapenem Regimen in Pandrug-Resistant Klebsiella pneumoniae Bloodstream Infections. J. Antimicrob. Chemother. 2014, 69, 1718–1720.
- de Souza, G.H.d.A.; dos Santos Radai, J.A.; Mattos Vaz, M.S.; Esther da Silva, K.; Fraga, T.L.; Barbosa, L.S.; Simionatto, S. In Vitro and in Vivo Antibacterial Activity Assays of Carvacrol: A Candidate for Development of Innovative Treatments against KPC-Producing Klebsiella pneumoniae. PLoS ONE 2021, 16, e0246003.
- Qian, W.; Sun, Z.; Wang, T.; Yang, M.; Liu, M.; Zhang, J.; Li, Y. Antimicrobial Activity of Eugenol against Carbapenem-Resistant Klebsiella pneumoniae and Its Effect on Biofilms. Microb. Pathog. 2020, 139, 103924.
- Dhara, L.; Tripathi, A. Sub-Acute Toxicological and Behavioural Effects of Two Candidate Therapeutics, Cinnamaldehyde and Eugenol, for Treatment of ESBL Producing-Quinolone Resistant Pathogenic Enterobacteriaceae. Clin. Exp. Pharmacol. Physiol. 2020, 47, 977–988.
- Anwar, R.; Sukmasari, S.; Siti Aisyah, L.; Puspita Lestari, F.; Ilfani, D.; Febriani Yun, Y.; Diki Prestya, P. Antimicrobial Activity of β-Sitosterol Isolated from Kalanchoe tomentosa Leaves Against Staphylococcus aureus and Klebsiella pneumoniae. Pak. J. Biol. Sci. 2022, 25, 602–607.
- Al-Qahtani, W.H.; Dinakarkumar, Y.; Arokiyaraj, S.; Saravanakumar, V.; Rajabathar, J.R.; Arjun, K.; Gayathri, P.K.; Nelson Appaturi, J. Phyto-Chemical and Biological Activity of Myristica fragrans, an Ayurvedic Medicinal Plant in Southern India and Its Ingredient Analysis. Saudi J. Biol. Sci. 2022, 29, 3815–3821.
- Yang, S.-K.; Yusoff, K.; Ajat, M.; Thomas, W.; Abushelaibi, A.; Akseer, R.; Lim, S.-H.E.; Lai, K.-S. Disruption of KPC-Producing Klebsiella pneumoniae Membrane via Induction of Oxidative Stress by Cinnamon Bark (Cinnamomum verum J. Presl) Essential Oil. PLoS ONE 2019, 14, e0214326.
- Gopu, V.; Kothandapani, S.; Shetty, P.H. Quorum Quenching Activity of Syzygium cumini (L.) Skeels and Its Anthocyanin Malvidin against Klebsiella pneumoniae. Microb. Pathog. 2015, 79, 61–69.
- Supardy, N.A. Inhibition of Klebsiella pneumoniae ATCC 13883 Cells by Hexane Extract of Halimeda discoidea (Decaisne) and the Identification of Its Potential Bioactive Compounds. J. Microbiol. Biotechnol. 2012, 22, 872–881.
- Akbari, R.; Bafghi, M.F.; Fazeli, H. Nosocomial Infections Pathogens Isolated from Hospital Personnel, Hospital Environment and Devices. J. Med. Bacteriol. 2018, 7, 22–30.
- Zielińska, A.; Carreiró, F.; Oliveira, A.M.; Neves, A.; Pires, B.; Venkatesh, D.N.; Durazzo, A.; Lucarini, M.; Eder, P.; Silva, A.M.; et al. Polymeric Nanoparticles: Production, Characterization, Toxicology and Ecotoxicology. Molecules 2020, 25, 3731.
- Bharathala, S.; Sharma, P. Biomedical Applications of Nanoparticles. In Nanotechnology in Modern Animal Biotechnology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 113–132.
- Ying, S.; Guan, Z.; Ofoegbu, P.C.; Clubb, P.; Rico, C.; He, F.; Hong, J. Green Synthesis of Nanoparticles: Current Developments and Limitations. Environ. Technol. Innov. 2022, 26, 102336.
- Salomoni, R.; Léo, P.; Montemor, A.F.; Rinaldi, B.G.; Rodrigues, M.F.A. Antibacterial Effect of Silver Nanoparticles in Pseudomonas aeruginosa. Nanotechnol. Sci. Appl. 2017, 10, 115–121.
- Chinnathambi, A.; Alharbi, S.A.; Joshi, D.; Saranya, V.; Jhanani, G.K.; On-Uma, R.; Jutamas, K.; Anupong, W. Synthesis of AgNPs from Leaf Extract of Naringi crenulata and Evaluation of Its Antibacterial Activity against Multidrug Resistant Bacteria. Environ. Res. 2022, 216, 114455.
- Kabeerdass, N.; Murugesan, K.; Arumugam, N.; Almansour, A.I.; Kumar, R.S.; Djearamane, S.; Kumaravel, A.K.; Velmurugan, P.; Mohanavel, V.; Kumar, S.S.; et al. Biomedical and Textile Applications of Alternanthera sessilis Leaf Extract Mediated Synthesis of Colloidal Silver Nanoparticle. Nanomaterials 2022, 12, 2759.
- Wali, S.; Zahra, M.; Okla, M.K.; Wahidah, H.A.; Tauseef, I.; Haleem, K.S.; Farid, A.; Maryam, A.; AbdElgawad, H.; Adetunji, C.O.; et al. Brassica oleracea L. (Acephala Group) Based Zinc Oxide Nanoparticles and Their Efficacy as Antibacterial Agent. Braz. J. Biol. 2022, 84, e259351.
- Yuan, Y.; Wu, Y.; Suganthy, N.; Shanmugam, S.; Brindhadevi, K.; Sabour, A.; Alshiekheid, M.; Lan Chi, N.T.; Pugazhendhi, A.; Shanmuganathan, R. Biosynthesis of Zirconium Nanoparticles (ZrO(2) NPs) by Phyllanthus niruri Extract: Characterization and Its Photocatalytic Dye Degradation Activity. Food Chem. Toxicol. 2022, 168, 113340.
- Zhou, Y.; Xu, M.; Liu, Y.; Bai, Y.; Deng, Y.; Liu, J.; Chen, L. Green Synthesis of Se/Ru Alloy Nanoparticles Using Gallic Acid and Evaluation of Theiranti-Invasive Effects in HeLa Cells. Colloids Surf. B Biointerfaces 2016, 144, 118–124.
- Hassan, H.U.; Raja, N.I.; Abasi, F.; Mehmood, A.; Qureshi, R.; Manzoor, Z.; Shahbaz, M.; Proćków, J. Comparative Study of Antimicrobial and Antioxidant Potential of Olea ferruginea Fruit Extract and Its Mediated Selenium Nanoparticles. Molecules 2022, 27, 5194.
- Todorova, V.; Ivanov, K.; Georgieva, Y.; Karcheva-Bahchevanska, D.; Ivanova, S. Comparison between the Chemical Composition of Essential Oil from Commercial Products and Biocultivated Lavandula angustifolia Mill. Int. J. Anal. Chem. 2023, 2023, 1997157.
- Chung, W.-Y.; Jadhav, S.; Hsu, P.-K.; Kuan, C.-M. Evaluation of Acute and Sub-Chronic Toxicity of Bitter Melon Seed Extract in Wistar Rats. Toxicol. Rep. 2022, 9, 1024–1034.
- Sireeratawong, S.; Jaijoy, K.; Khonsung, P.; Lertprasertsuk, N.; Ingkaninan, K. Acute and Chronic Toxicities of Bacopa monnieri Extract in Sprague-Dawley Rats. BMC Complement Altern. Med. 2016, 16, 249.
- Ahmad, F.; Tabassum, N. Preliminary Phytochemical, Acute Oral Toxicity and Antihepatotoxic Study of Roots of Paeonia officinalis Linn. Asian Pac. J. Trop. Biomed. 2013, 3, 64–68.
- Sulaiman, C.T.; Gopalakrishnan, V.K. Radical Scavenging and In-Vitro Hemolytic Activity of Aqueous Extracts of Selected Acacia Species. J. Appl. Pharm. Sci. 2013, 3, 109–111.
- Liaqat, I.; Mahreen, A.; Arshad, M.; Arshad, N. Antimicrobial and Toxicological Evaluation of Origanum vulgare: An in Vivo Study. Braz. J. Biol. 2023, 83, e244551.
- Rojas-Armas, J.; Arroyo-Acevedo, J.; Ortiz-Snchez, M.; Palomino-Pacheco, M.; Castro-Luna, A.; Ramos-Cevallos, N.; Justil-Guerrero, H.; Hilario-Vargas, J.; Herrera-Caldern, O. Acute and Repeated 28-Day Oral Dose Toxicity Studies of Thymus vulgaris L. Essential Oil in Rats. Toxicol. Res. 2019, 35, 225–232.
- Prashar, A.; Locke, I.C.; Evans, C.S. Cytotoxicity of Clove (Syzygium aromaticum) Oil and Its Major Components to Human Skin Cells. Cell Prolif. 2006, 39, 241–248.
- Wijesinghe, G.K.; Maia, F.C.; de Oliveira, T.R.; de Feiria, S.N.B.; Joia, F.; Barbosa, J.P.; Boni, G.C.; de Cássia Orlandi Sardi, J.; Rosalen, P.L.; Höfling, J.F. Effect of Cinnamomum verum Leaf Essential Oil on Virulence Factors of Candida Species and Determination of the In-Vivo Toxicity with Galleria Mellonella Model. Mem Inst Oswaldo Cruz 2020, 115.
- Nair, B. Final Report on the Safety Assessment of Mentha piperita (Peppermint) Oil, Mentha piperita (Peppermint) Leaf Extract, Mentha piperita (Peppermint) Leaf, and Mentha piperita (Peppermint) Leaf Water. Int. J. Toxicol. 2001, 20 (Suppl. S3), 61–73.
- Fahim, F.; Esmat, A.; Fadel, H.; Hassan, K. Allied Studies on the Effect of Rosmarinus officinalis L. on Experimental Hepatotoxicity and Mutagenesis. Int. J. Food Sci. Nutr. 1999, 50, 413–427.
- Aggarwal, M.L.; Chacko, K.M.; Kuruvilla, B.T. Systematic and Comprehensive Investigation of the Toxicity of Curcuminoid-Essential Oil Complex: A Bioavailable Turmeric Formulation. Mol. Med. Rep. 2016, 13, 592–604.
More
