Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Mona Zou and Version 1 by Gloria Borgstahl.
Human Replication Protein A (RPA) was historically discovered as one of the six components needed to reconstitute simian virus 40 DNA replication from purified components. RPA is now known to be involved in all DNA metabolism pathways that involve single-stranded DNA (ssDNA). Heterotrimeric RPA comprises several domains connected by flexible linkers and is heavily regulated by post-translational modifications (PTMs). The structure of RPA has been challenging to obtain. Various structural methods have been applied, but a complete understanding of RPA’s flexible structure, its function, and how it is regulated by PTMs has yet to be obtained.
- Replication Protein A (RPA)
- phosphorylation
- homologous recombination
- AlphaFold
- protein-ssDNA interactions
- cell cycle
- DNA metabolism
- double-strand break repair
1. The Function of RPA in DNA Double-Strand Break Repair
RPA is critical in coordinating the cell cycle with DNA replication and any DNA repair pathway involving ssDNA. The cell cycle is crucial in completing a series of events that allow cells to grow and divide and includes the following stages: interphase (I), G1 phase, S phase, G2 phase, and mitosis (M). Regulation is key to successfully replicating DNA and normal cell division within these cell cycle phases. Several laboratories have studied RPA in cell cycle regulation. RPA was determined not to be phosphorylated in the G1 phase, but RPA34 in humans and S. cerevisiae is phosphorylated in cells that enter the S phase and then are dephosphorylated in the M phase [45,87,88][1][2][3]. Various researchers have focused on RPA’s role in recruiting proteins involved in DNA repair. RPA is involved in most DNA repair pathways, including base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), and HR [36,89,90,91,92][4][5][6][7][8]. It is also important to note that HR occurs only during S and G2 phases [40,44][9][10].
RPA plays a critical role in HR to repair DSBs. In HR, sections of homologous sequences, typically on a sister chromatid or nearby repeated sequence, will fill in the gap left by the DSB. On each side of this DSB, a 5′ strand is resected, leaving a 3′ overhang on the DNA. This resection is done by the MRN complex (composed of RAD50, mitotic recombination 11 (MRE11), and Nijmegen breakage syndrome 1 (NBS1)), along with other proteins, including CtBP-interacting protein (CtIP), exonuclease 1 (EXO1), DNA replication helicase/nuclease DNA2, and blood syndrome RecQ like helicase (BLM). Once there is a 3′ ssDNA overhang, RPA will bind to this ssDNA and follow the canonical HR pathway that will recruit a complex that includes breast cancer type 1 susceptibility protein (BRCA1), breast cancer type 2 susceptibility protein (BRCA2), and partner and localizer of BRCA2 (PALB1). This BRCA2 complex will recruit radiation sensitivity gene 51 (RAD51) and undergo ssDNA transfer from RPA to BRCA2 complex to RAD51, and RAD51 will perform a homology search on the sister chromatid, eventually repairing the DSB. Secondly, an alternative pathway for HR includes radiation sensitivity gene 52 (RAD52). RAD52 has similar functionality to the BRCA2 complex by undergoing a ssDNA handoff from RPA and allowing RAD51 nucleation of the ssDNA again to perform a homology search on the sister chromatid, eventually repairing the DSB [40,53,93,94,95][9][11][12][13][14].
2. ssDNA and Protein Interactions involving RPA
RPA involves an assorted array of interactions with ssDNA and proteins. The function of RPA binding to ssDNA has been studied in numerous ways, for example, through atomic force microscopy (AFM), circular dichroism (CD), optical tweezers, single-molecule fluorescence resonance energy transfer (smFRET), and many more [61,96,97][15][16][17]. The interaction between ssDNA and RPA is a rapid but extremely stable process that is difficult to evaluate due to the binding strength. A study in 2000 concluded that the zinc-finger motif is necessary to form a stable RPA:ssDNA complex, which depends on redox reactions [98][18]. Others focused on the binding and unfolding of non-canonical ssDNA structures by RPA in DNA replication. The heterotrimeric core of RPA selectively binds to a G-quadruplex forming sequence. G-quadruplexes create a unique problem for RPA to unfold, and various ligands could hinder DNA replication by not allowing RPA to unfold these G-quadruplexes correctly [99][19].
A more recent paper described how the ssDNA binding complex can change its binding mode to allow for nucleation of RAD51. The spacing between RPA:ssDNA complexes is essential for leaving bare nucleotides in between the complexes to allow for RAD51 nucleation. The exchange of RPA for RAD51 is known to be mediated by the RAD52 middle region [100][20]. Overall, the RPA:ssDNA complex is essential for life and continues to be investigated.
With this insight into the RPA:ssDNA complex, multiple laboratories have investigated how RPA binds to varying lengths of nucleotides (nt), for example, 8-nt, 20-nt, or 30-nt. Kang and coworkers in 2023 investigated a protein involved in Kallmann syndrome: N-methyl-D-aspartate receptor synaptonuclear signaling and neuronal migration factor (NSMF). They determined that NSMF will colocalize and physically interact with RPA during DNA damage, allowing RPA to bind in a 30-nt binding mode. This prepares RPA to become phosphorylated by ATR, an upstream kinase involved in HR [101][21]. They concluded that the 30-nt binding mode of RPA enhances the phosphorylation of RPA32 by ATR, and further phosphorylation will stabilize RPA binding to ssDNA.
RPA has many protein–protein interactions involving DSB repair proteins and others (Table 21). DSB repair binding partners include: BRCA2, RAD52, RAD51, ATR/ATRIP, DNA-PKcs, DSS1, MRE11-RAD50-NBS1, p53, and PP2A [18,21,36,38,69,72,74,90,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212][4][6][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73][74][75][76][77][78][79][80][81][82][83][84][85][86][87][88][89][90][91][92][93][94][95][96][97][98][99][100][101][102][103][104][105][106][107][108][109][110][111][112][113][114][115][116][117][118][119][120][121][122][123][124][125][126][127][128][129][130][131][132][133][134][135][136][137][138]. The interaction of RPA with RAD52 in the alternative HR DSB repair pathway and single-strand annealing is fascinating. Without the RAD52:RPA interaction, the ssDNA handoff from RPA to RAD51 in alternative HR would not be possible, and it is also critical that RPA is phosphorylated to allow this DNA handoff [93][12]. Previous studies identified residues 224-271 on RPA32 and 169-326 on RPA70 that include binding sites for RAD52. It was also found that RAD52 residues 218-303 bind RPA70, along with RPA32 [21][23]. Although the interaction sites have been defined, structural data has yet to be available for the RPA:RAD52 complex.
Table 21. Human proteins that interact with RPA.
| Interacting Protein | Interaction Site on RPA | DNA Metabolism Pathway | Citation |
|---|---|---|---|
| AID | RPA32 | Immunoglobulin diversification | [107][33] |
| * Ajuba | RPA70 | DNA damage response (DDR) | [108,109][34][35] |
| ** ATR/ATRIP | RPA70-F | Checkpoint signaling, DNA repair | [110,111,112][36][37][38] |
| BID | RPA70-F | Replication stress response | [113][39] |
| * BLM | RPA70-A/B | DNA unwinding, resection | [114,115,125][40][41][51] |
| ** BRCA2 | ? | Homologous Recombination (HR) | [105][31] |
| DDX11 | ? | Chromosome segregation | [193][119] |
| ** DNA-PKcs | ? | DNA repair | [18,116][22][42] |
| ** DSS1 | RPA70 | HR | [121][47] |
| * ETAA1 | RPA70-F/RPA32 | ATR activation, repair at stalled replication forks |
[117,118,119,120][43][44][45][46] |
| * EXO5 | RPA70-F | Intrastrand crosslink repair | [181][107] |
| * FANCJ | RPA70 | DNA repair, genome stability | [123][49] |
| * FBH1 | RPA32 | DNA unwinding, resection | [193][119] |
| HELB | RPA70-F | Replication stress response | [179,180][105][106] |
| HERC2 | RPA70 | Replication | [124,125][50][51] |
| HIRA | RPA70-C | Chromatin remodeling | [126][52] |
| Histones H3 and H4 | RPA70-F | Chromatin remodeling | [127][53] |
| * HLTF | RPA70 | Genome stability | [195,196][121][122] |
| HSF1 | RPA70 | Gene expression | [122][48] |
| * Menin | RPA32 | Genome stability | [129,130][55][56] |
| ** MRE11-RAD50-NBS1 | RPA70-F | HR | [131,132][57][58] |
| Nucleolin | RPA14 | Replication (stress) | [133,134,135][59][60][61] |
| NSMF | RPA32 | DDR | [101][21] |
| ** p53 | RPA70-F | HR | [136,137,138,139,140][62][63][64][65][66] |
| * p53BP1 | RPA70/RPA32 | DDR | [106][32] |
| * PALB2 | RPA32 | Recovery of stalled replication forks | [141][67] |
| Papillomavirus E1 | RPA70-A | Replication | [182,183][108][109] |
| Parvovirus NS1 | RPA70/RPA32 | DNA unwinding, resection | [187][113] |
| PCNA | RPA70 | Replication | [191][117] |
| Polδ | RPA70 | Replication | [188][114] |
| Pol-Prim | RPA70-F/A/B | Replication restart, DNA damage tolerance |
[102,203][28][129] |
| ** PP2A | RPA32 | DDR | [204][130] |
| * PRP19/BCAS2 | RPA70-F/C | DNA repair | [144][70] |
| * PTEN | RPA70 | Genome stability | [145][71] |
| * RAD9/RAD1/HUS1 (9-1-1) | RPA70/RPA32 | DDR | [146][72] |
| RAD17 | RPA70-F | DDR, replication stress response | [147,178,205][73][104][131] |
| ** RAD51 | RPA70-A | Recombination | [103,150,151,206][29][76][77][132] |
| ** RAD52 | RPA70-A/B & RPA32-wHLH | DNA repair | [21,69,152,][23]153,[25]154,[155,156,78][79][80][81][82]207[133] |
| RECQL1 | RPA70 | DNA unwinding | [198,199][124][125] |
| RECQ5β | ? | DNA unwinding | [200,201][126][127] |
| RFC | RPA70 | DNA unwinding | [188,189][114][115] |
| * RFWD3 | RPA32-wHLH | DNA repair | [157,158,195][83][84][121] |
| * RNaseH | ? | Transcription, DNA repair | [159][85] |
| * RNF4 | RPA70 | DNA DSB repair by HR | [202][128] |
| SENP6 | RPA70 | Unperturbed DNA replication | [38][24] |
| SMARCAL1/HARP | RPA32-wHLH | Replication fork restart | [160,161,162,208][86][87][88][134] |
| SV40 Large T antigen | RPA70-A/B & RPA32-wHLH | Replication | [182,183,184,185,186][108][109][110][111][112] |
| * Tipin | RPA32-wHLH | DDR | [164,165][90][91] |
| * UDG | RPA32-wHLH | Base excision repair | [69,166,190][25][92][116] |
| * UNG2 | RPA32-wHLH | Base excision repair | [69,166][25][92] |
| * WRN | RPA70-A/B | DNA unwinding, resection | [115,170][41][96] |
| * XPA | RPA70-A & RPA32-wHLH | Nucleotide excision repair (NER) | [69,166,171,209,210,211][25][92][97][135][136][137] |
| * XPF-ERCC1 | RPA70 | NER | [174,175,176][100][101][102] |
| * XPG | ? | NER | [174,176,212][100][102][138] |
* Involved in DNA repair; ** Involved in DNA DSB repair.
BRCA2 is an essential protein involved in the canonical HR pathway. BRCA2 also completes the DNA handoff, like RAD52, by interacting with RPA. BRCA2 mutations in women can cause familial, early-onset breast cancer, so understanding these mutations and their potential impact on the interaction with RPA will become vital in understanding DNA repair, specifically in cancer cells. A study in 2003 used a cancer-predisposing BRCA2 mutation (Y42C) to investigate its interaction with RPA. They found that the Y42C mutation inhibited the interaction between RPA:BRCA2, showing that the Y42C mutation has biological importance within the human body [213][139]. The BRCA2 interaction with RAD51 has been studied extensively over the years, as BRCA2 promotes RAD51 nucleation on the ssDNA. Still, there has yet to be data on the interaction between BRCA2 and RPA to specifically understand how and where this interaction occurs [214][140].
3. PTMs That Regulate RPA in DNA Metabolism
PTMs are chemical modifications involved in the functional regulation of cellular proteins. RPA relies on PTMs for its proper function in the cell cycle and DNA repair, and the main PTM will be focused here is phosphorylation. Phosphorylation of RPA has been studied for decades, with most studies focused on the RPA32 N-terminal region. Understanding the importance of phosphorylation on other RPA subunits in RPA’s regulation is still unresolved.
Phosphorylation occurs throughout the cell cycle, as previously described. A paper by Yates and coworkers determined two cell cycle checkpoint kinases in yeast, Mec1 and Ddc2 (ATR and ATRIP, respectively, in humans), that are essential for replication stress response and DDR. These two kinases were shown to recruit ssDNA binding to RPA through Ddc2 by phosphorylation. A yeast Ddc2-RPA structure through X-ray crystallography indicated that this interaction is necessary to mediate RPA phosphorylation during DDR [215][141].
An early study in 1990 by Din and colleagues determined the phosphorylation of RPA32 and RPA70 in human and yeast cells by phosphoamino acid analysis using P32 labeling [39][142]. Then, numerous labs focused on the phosphorylation of RPA because of its critical regulation in the DNA repair pathway. In 2003, Binz and others discovered that the phosphorylation of RPA32 modulates RPA-dsDNA interactions and subsequent destabilization [22][143]. They concluded that an intersubunit interaction between phosphorylated RPA32NT and RPA70 N-terminal domain (RPA70N) was possible. Recently, NMR and docking studies were conducted to investigate the interaction between the RPA70 N-terminal domain (RPA70N) and a phosphomimetic N-terminal peptide of RPA32 with candidate serine/threonine sites mutated to aspartic acid. The study showed the possibility that RPA32 N-terminal phosphorylation could allow for transient interaction with RPA70N, which could alter or enhance other interaction sites for proteins or DNA [216][144]. These conclusions indicated a possible role of RPA32 N-terminal phosphorylation binding with RPA70N to form a less disordered structure if these interactions occur in the context of the heterotrimer.
Not only is RPA involved in RPA-DNA interactions, but RPA phosphorylation also influences subcellular localization, especially when there is mitotic phosphorylation at S23 and S29 in RPA32 [217][145]. When the cell has a DDR, RPA becomes hyperphosphorylated [50,56][146][147]. For example, in response to DSBs, RPA is a substrate of ATM kinase that phosphorylates RPA32 at Thr21 and most likely others [43][148]. In 2014, a study was performed on RPA hyperphosphorylation involving the HR pathway in cell cycle phases where HR DSB repair occurs. In these studies, a human squamous cell carcinoma cell line (UM-SCC-38) was synchronized in the S and G2 phases [218,219,220][149][150][151]. Phosphorylation sites in S and G2 phases with and without DSBs were analyzed with western blots using all available antibodies for phosphorylation sites on RPA32. In the G2 phase, chromatin-bound immunoprecipitation phosphorylation occurred at S4/8, S12, T21, S23, and S33 of RPA32 after DSBs.
A global view of RPA heterotrimer phosphorylation was explained using a high-quality RPA70 antibody and isoelectric focusing (IEF) to separate RPA isoforms based on the number of phosphorylations present. Capillary IEF immunoassay data of the phosphorylated RPA heterotrimeric isoforms with and without DSB treatment, synchronized in the G2 phase of the cell cycle, were collected (Figure 21) [40][9]. In this study, data showed that RPA is always a phosphoprotein in the G2 phase by having up to seven phosphates (Figure 21A, blue line). Hyperphosphorylated isoforms generated after DSBs contained up to 13 or 14 phosphates. IEF identified some of these sites with phosphospecific antibodies (Figure 21B–E). The hyperphosphorylated forms with 13–14 phosphates included pS4/8, pT21, and pS33. Several of the sites had yet to be characterized before. DSBs showed increased phosphorylation by PIKK and CDK kinases in S and G2 phases, providing new information on candidate PIKK and CDK sites for future research.
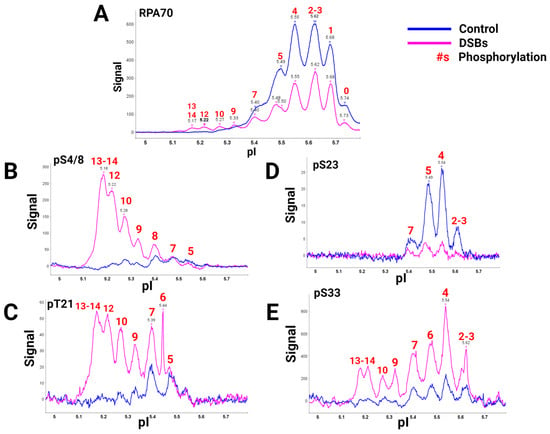
Figure 21. RPA isoforms from cells synced in the G2 phase either with DNA damage (blue) or without DNA damage (pink), separated with isoelectric focusing using a pH gradient from 5-6 and probed with phospho-specific antibodies. RPA heterotrimer probed with (A) anti-RPA70-CT, (B) phosphoRPA32(S4/8), (C) phosphoRPA32(T21), (D) phosphoRPA32(S23), and (E) phosphoRPA32(S33) antibodies. The red number above the corresponding peaks indicates the number of phosphorylations on each isoform. Figure adapted from [40][9].
This research provided pertinent information about RPA heterotrimeric phosphorylation in response to DSBs in the S and G2 phases of the human cell cycle. Several of these phosphorylation sites are PIKK-specific sites in the DNA binding and protein interaction domains of RPA. A summary figure describes possible phosphorylation sites before and after DSBs (Figure 32). This study provided data about phosphatase and kinase action in remodeling RPA isoforms in response to DNA damage [40][9].
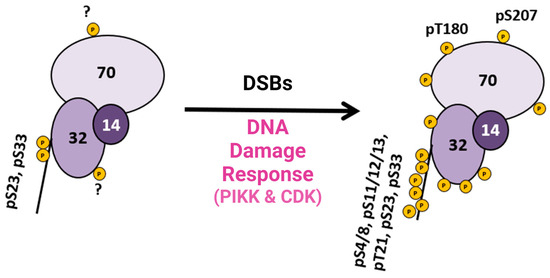
Figure 32. Summary diagram of RPA phosphorylation before and after DSBs. RPA is always phosphorylated (some sites are unknown and indicated with a ?), but after DDR is activated, RPA becomes hyperphosphorylated by the PIKK family of kinases and CDK cell cycle kinases, which phosphorylate RPA. Phosphorylation sites after DNA damage are shown in Figure 1D. Adapted from [40][9].
References
- Stillman, B.; Bell, S.P.; Dutta, A.; Marahrens, Y. DNA replication and the cell cycle. Ciba Found. Symp. 1992, 170, 147–156; discussion 156–160.
- Dutta, A.; Din, S.; Brill, S.J.; Stillman, B. Phosphorylation of replication protein A: A role for cdc2 kinase in G1/S regulation. Cold Spring Harb. Symp. Quant. Biol. 1991, 56, 315–324.
- Murti, K.G.; He, D.C.; Brinkley, B.R.; Scott, R.; Lee, S.H. Dynamics of human replication protein A subunit distribution and partitioning in the cell cycle. Exp. Cell Res. 1996, 223, 279–289.
- He, H.; Wang, J.; Liu, T. UV-Induced RPA1 Acetylation Promotes Nucleotide Excision Repair. Cell Rep. 2017, 20, 2010–2025.
- Thoma, B.S.; Vasquez, K.M. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol. Carcinog. 2003, 38, 1–13.
- Zhao, M.; Geng, R.; Guo, X.; Yuan, R.; Zhou, X.; Zhong, Y.; Huo, Y.; Zhou, M.; Shen, Q.; Li, Y.; et al. PCAF/GCN5-Mediated Acetylation of RPA1 Promotes Nucleotide Excision Repair. Cell Rep. 2017, 20, 1997–2009.
- Hayran, A.B.; Liabakk, N.B.; Aas, P.A.; Kusnierczyk, A.; Vagbo, C.B.; Sarno, A.; Iveland, T.S.; Chawla, K.; Zahn, A.; Di Noia, J.M.; et al. RPA guides UNG to uracil in ssDNA to facilitate antibody class switching and repair of mutagenic uracil at the replication fork. Nucleic Acids Res. 2024, 52, 784–800.
- Gavande, N.S.; VanderVere-Carozza, P.S.; Hinshaw, H.D.; Jalal, S.I.; Sears, C.R.; Pawelczak, K.S.; Turchi, J.J. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol. Ther. 2016, 160, 65–83.
- Borgstahl, G.E.; Brader, K.; Mosel, A.; Liu, S.; Kremmer, E.; Goettsch, K.A.; Kolar, C.; Nasheuer, H.P.; Oakley, G.G. Interplay of DNA damage and cell cycle signaling at the level of human replication protein A. DNA Repair 2014, 21, 12–23.
- Priya, B.; Ravi, S.; Kirubakaran, S. Targeting ATM and ATR for cancer therapeutics: Inhibitors in clinic. Drug Discov. Today 2023, 28, 103662.
- Prakash, A.; Borgstahl, G.E. The structure and function of replication protein A in DNA replication. Subcell. Biochem. 2012, 62, 171–196.
- Deng, X.; Prakash, A.; Dhar, K.; Baia, G.S.; Kolar, C.; Oakley, G.G.; Borgstahl, G.E. Human replication protein A-Rad52-single-stranded DNA complex: Stoichiometry and evidence for strand transfer regulation by phosphorylation. Biochemistry 2009, 48, 6633–6643.
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018, 293, 10524–10535.
- Creeden, J.F.; Nanavaty, N.S.; Einloth, K.R.; Gillman, C.E.; Stanbery, L.; Hamouda, D.M.; Dworkin, L.; Nemunaitis, J. Homologous recombination proficiency in ovarian and breast cancer patients. BMC Cancer 2021, 21, 1154.
- Kusi-Appauh, N.; Ralph, S.F.; van Oijen, A.M.; Spenkelink, L.M. Understanding G-Quadruplex Biology and Stability Using Single-Molecule Techniques. J. Phys. Chem. B 2023, 127, 5521–5540.
- Prakash, A.; Natarajan, A.; Marky, L.A.; Ouellette, M.M.; Borgstahl, G.E. Identification of the DNA-Binding Domains of Human Replication Protein A That Recognize G-Quadruplex DNA. J. Nucleic Acids 2011, 2011, 896947.
- Lysetska, M.; Knoll, A.; Boehringer, D.; Hey, T.; Krauss, G.; Krausch, G. UV light-damaged DNA and its interaction with human replication protein A: An atomic force microscopy study. Nucleic Acids Res. 2002, 30, 2686–2691.
- You, J.S.; Wang, M.; Lee, S.H. Functional characterization of zinc-finger motif in redox regulation of RPA-ssDNA interaction. Biochemistry 2000, 39, 12953–12958.
- Prakash, A.; Kieken, F.; Marky, L.A.; Borgstahl, G.E. Stabilization of a G-Quadruplex from Unfolding by Replication Protein A Using Potassium and the Porphyrin TMPyP4. J. Nucleic Acids 2011, 2011, 529828.
- Ding, J.; Li, X.; Shen, J.; Zhao, Y.; Zhong, S.; Lai, L.; Niu, H.; Qi, Z. ssDNA accessibility of Rad51 is regulated by orchestrating multiple RPA dynamics. Nat. Commun. 2023, 14, 3864.
- Kang, Y.; Han, Y.G.; Khim, K.W.; Choi, W.G.; Ju, M.K.; Park, K.; Shin, K.J.; Chae, Y.C.; Choi, J.H.; Kim, H.; et al. Alteration of replication protein A binding mode on single-stranded DNA by NSMF potentiates RPA phosphorylation by ATR kinase. Nucleic Acids Res. 2023, 51, 7936–7950.
- Shao, R.G.; Cao, C.X.; Zhang, H.; Kohn, K.W.; Wold, M.S.; Pommier, Y. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999, 18, 1397–1406.
- Jackson, D.; Dhar, K.; Wahl, J.K.; Wold, M.S.; Borgstahl, G.E. Analysis of the human replication protein A:Rad52 complex: Evidence for crosstalk between RPA32, RPA70, Rad52 and DNA. J. Mol. Biol. 2002, 321, 133–148.
- Dou, H.; Huang, C.; Singh, M.; Carpenter, P.B.; Yeh, E.T. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 2010, 39, 333–345.
- Mer, G.; Bochkarev, A.; Gupta, R.; Bochkareva, E.; Frappier, L.; Ingles, C.J.; Edwards, A.M.; Chazin, W.J. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 2000, 103, 449–456.
- Bochkareva, E.; Kaustov, L.; Ayed, A.; Yi, G.S.; Lu, Y.; Pineda-Lucena, A.; Liao, J.C.; Okorokov, A.L.; Milner, J.; Arrowsmith, C.H.; et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. USA 2005, 102, 15412–15417.
- Guilliam, T.A.; Brissett, N.C.; Ehlinger, A.; Keen, B.A.; Kolesar, P.; Taylor, E.M.; Bailey, L.J.; Lindsay, H.D.; Chazin, W.J.; Doherty, A.J. Molecular basis for PrimPol recruitment to replication forks by RPA. Nat. Commun. 2017, 8, 15222.
- Braun, K.A.; Lao, Y.; He, Z.; Ingles, C.J.; Wold, M.S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase Alpha by multiple mechanisms. Biochemistry 1997, 36, 8443–8454.
- Golub, E.I.; Gupta, R.C.; Haaf, T.; Wold, M.S.; Radding, C.M. Interaction of human rad51 recombination protein with single-stranded DNA binding protein, RPA. Nucleic Acids Res. 1998, 26, 5388–5393.
- Riva, F.; Zuco, V.; Vink, A.A.; Supino, R.; Prosperi, E. UV-induced DNA incision and proliferating cell nuclear antigen recruitment to repair sites occur independently of p53-replication protein A interaction in p53 wild type and mutant ovarian carcinoma cells. Carcinogenesis 2001, 22, 1971–1978.
- Wang, M.; Mahrenholz, A.; Lee, S.H. RPA stabilizes the XPA-damaged DNA complex through protein-protein interaction. Biochemistry 2000, 39, 6433–6439.
- Yoo, E.; Kim, B.U.; Lee, S.Y.; Cho, C.H.; Chung, J.H.; Lee, C.H. 53BP1 is associated with replication protein A and is required for RPA2 hyperphosphorylation following DNA damage. Oncogene 2005, 24, 5423–5430.
- Chaudhuri, J.; Khuong, C.; Alt, F.W. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature 2004, 430, 992–998.
- Kalan, S.; Matveyenko, A.; Loayza, D. LIM Protein Ajuba Participates in the Repression of the ATR-Mediated DNA Damage Response. Front. Genet. 2013, 4, 95.
- Fowler, S.; Maguin, P.; Kalan, S.; Loayza, D. LIM Protein Ajuba associates with the RPA complex through direct cell cycle-dependent interaction with the RPA70 subunit. Sci. Rep. 2018, 8, 9536.
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548.
- Ball, H.L.; Myers, J.S.; Cortez, D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell 2005, 16, 2372–2381.
- Namiki, Y.; Zou, L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc. Natl. Acad. Sci. USA 2006, 103, 580–585.
- Liu, Y.; Vaithiyalingam, S.; Shi, Q.; Chazin, W.J.; Zinkel, S.S. BID binds to replication protein A and stimulates ATR function following replicative stress. Mol. Cell Biol. 2011, 31, 4298–4309.
- Brosh, R.M., Jr.; Li, J.L.; Kenny, M.K.; Karow, J.K.; Cooper, M.P.; Kureekattil, R.P.; Hickson, I.D.; Bohr, V.A. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000, 275, 23500–23508.
- Doherty, K.M.; Sommers, J.A.; Gray, M.D.; Lee, J.W.; von Kobbe, C.; Thoma, N.H.; Kureekattil, R.P.; Kenny, M.K.; Brosh, R.M., Jr. Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J. Biol. Chem. 2005, 280, 29494–29505.
- Blackwell, L.J.; Borowiec, J.A.; Mastrangelo, I.A. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell Biol. 1996, 16, 4798–4807.
- Feng, S.; Zhao, Y.; Xu, Y.; Ning, S.; Huo, W.; Hou, M.; Gao, G.; Ji, J.; Guo, R.; Xu, D. Ewing Tumor-associated Antigen 1 Interacts with Replication Protein A to Promote Restart of Stalled Replication Forks. J. Biol. Chem. 2016, 291, 21956–21962.
- Haahr, P.; Hoffmann, S.; Tollenaere, M.A.; Ho, T.; Toledo, L.I.; Mann, M.; Bekker-Jensen, S.; Raschle, M.; Mailand, N. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 2016, 18, 1196–1207.
- Bass, T.E.; Luzwick, J.W.; Kavanaugh, G.; Carroll, C.; Dungrawala, H.; Glick, G.G.; Feldkamp, M.D.; Putney, R.; Chazin, W.J.; Cortez, D. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016, 18, 1185–1195.
- Lee, Y.C.; Zhou, Q.; Chen, J.; Yuan, J. RPA-Binding Protein ETAA1 Is an ATR Activator Involved in DNA Replication Stress Response. Curr. Biol. 2016, 26, 3257–3268.
- Zhao, W.; Vaithiyalingam, S.; San Filippo, J.; Maranon, D.G.; Jimenez-Sainz, J.; Fontenay, G.V.; Kwon, Y.; Leung, S.G.; Lu, L.; Jensen, R.B.; et al. Promotion of BRCA2-Dependent Homologous Recombination by DSS1 via RPA Targeting and DNA Mimicry. Mol. Cell 2015, 59, 176–187.
- Fujimoto, M.; Takaki, E.; Takii, R.; Tan, K.; Prakasam, R.; Hayashida, N.; Iemura, S.; Natsume, T.; Nakai, A. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol. Cell 2012, 48, 182–194.
- Gupta, R.; Sharma, S.; Sommers, J.A.; Kenny, M.K.; Cantor, S.B.; Brosh, R.M., Jr. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 2007, 110, 2390–2398.
- Lai, Y.; Zhu, M.; Wu, W.; Rokutanda, N.; Togashi, Y.; Liang, W.; Ohta, T. HERC2 regulates RPA2 by mediating ATR-induced Ser33 phosphorylation and ubiquitin-dependent degradation. Sci. Rep. 2019, 9, 14257.
- Wu, W.; Rokutanda, N.; Takeuchi, J.; Lai, Y.; Maruyama, R.; Togashi, Y.; Nishikawa, H.; Arai, N.; Miyoshi, Y.; Suzuki, N.; et al. HERC2 Facilitates BLM and WRN Helicase Complex Interaction with RPA to Suppress G-Quadruplex DNA. Cancer Res. 2018, 78, 6371–6385.
- Zhang, H.; Gan, H.; Wang, Z.; Lee, J.H.; Zhou, H.; Ordog, T.; Wold, M.S.; Ljungman, M.; Zhang, Z. RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol. Cell 2017, 65, 272–284.
- Liu, S.; Xu, Z.; Leng, H.; Zheng, P.; Yang, J.; Chen, K.; Feng, J.; Li, Q. RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. Science 2017, 355, 415–420.
- Dueva, R.; Iliakis, G. Replication protein A: A multifunctional protein with roles in DNA replication, repair and beyond. NAR Cancer 2020, 2, zcaa022.
- Sukhodolets, K.E.; Hickman, A.B.; Agarwal, S.K.; Sukhodolets, M.V.; Obungu, V.H.; Novotny, E.A.; Crabtree, J.S.; Chandrasekharappa, S.C.; Collins, F.S.; Spiegel, A.M.; et al. The 32-kilodalton subunit of replication protein A interacts with menin, the product of the MEN1 tumor suppressor gene. Mol. Cell Biol. 2003, 23, 493–509.
- Chen, C.C.; Juan, C.W.; Chen, K.Y.; Chang, Y.C.; Lee, J.C.; Chang, M.C. Upregulation of RPA2 promotes NF-kappaB activation in breast cancer by relieving the antagonistic function of menin on NF-kappaB-regulated transcription. Carcinogenesis 2017, 38, 196–206.
- Robison, J.G.; Elliott, J.; Dixon, K.; Oakley, G.G. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem. 2004, 279, 34802–34810.
- Oakley, G.G.; Tillison, K.; Opiyo, S.A.; Glanzer, J.G.; Horn, J.M.; Patrick, S.M. Physical interaction between replication protein A (RPA) and MRN: Involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry 2009, 48, 7473–7481.
- Daniely, Y.; Borowiec, J.A. Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol. 2000, 149, 799–810.
- Kim, K.; Dimitrova, D.D.; Carta, K.M.; Saxena, A.; Daras, M.; Borowiec, J.A. Novel checkpoint response to genotoxic stress mediated by nucleolin-replication protein a complex formation. Mol. Cell Biol. 2005, 25, 2463–2474.
- Wang, Y.; Guan, J.; Wang, H.; Wang, Y.; Leeper, D.; Iliakis, G. Regulation of DNA replication after heat shock by replication protein a-nucleolin interactions. J. Biol. Chem. 2001, 276, 20579–20588.
- Li, R.; Botchan, M.R. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 1993, 73, 1207–1221.
- He, Z.; Brinton, B.T.; Greenblatt, J.; Hassell, J.A.; Ingles, C.J. The transactivator proteins VP16 and GAL4 bind replication factor A. Cell 1993, 73, 1223–1232.
- Dutta, A.; Ruppert, J.M.; Aster, J.C.; Winchester, E. Inhibition of DNA replication factor RPA by p53. Nature 1993, 365, 79–82.
- Romanova, L.Y.; Willers, H.; Blagosklonny, M.V.; Powell, S.N. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene 2004, 23, 9025–9033.
- Serrano, M.A.; Li, Z.; Dangeti, M.; Musich, P.R.; Patrick, S.; Roginskaya, M.; Cartwright, B.; Zou, Y. DNA-PK, ATM and ATR collaboratively regulate p53-RPA interaction to facilitate homologous recombination DNA repair. Oncogene 2013, 32, 2452–2462.
- Murphy, A.K.; Fitzgerald, M.; Ro, T.; Kim, J.H.; Rabinowitsch, A.I.; Chowdhury, D.; Schildkraut, C.L.; Borowiec, J.A. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 2014, 206, 493–507.
- Wan, L.; Lou, J.; Xia, Y.; Su, B.; Liu, T.; Cui, J.; Sun, Y.; Lou, H.; Huang, J. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 2013, 14, 1104–1112.
- Guilliam, T.A.; Jozwiakowski, S.K.; Ehlinger, A.; Barnes, R.P.; Rudd, S.G.; Bailey, L.J.; Skehel, J.M.; Eckert, K.A.; Chazin, W.J.; Doherty, A.J. Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins. Nucleic Acids Res. 2015, 43, 1056–1068.
- Marechal, A.; Li, J.M.; Ji, X.Y.; Wu, C.S.; Yazinski, S.A.; Nguyen, H.D.; Liu, S.; Jimenez, A.E.; Jin, J.; Zou, L. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 2014, 53, 235–246.
- Wang, G.; Li, Y.; Wang, P.; Liang, H.; Cui, M.; Zhu, M.; Guo, L.; Su, Q.; Sun, Y.; McNutt, M.A.; et al. PTEN regulates RPA1 and protects DNA replication forks. Cell Res. 2015, 25, 1189–1204.
- Wu, X.; Shell, S.M.; Zou, Y. Interaction and colocalization of Rad9/Rad1/Hus1 checkpoint complex with replication protein A in human cells. Oncogene 2005, 24, 4728–4735.
- Zou, L.; Liu, D.; Elledge, S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 2003, 100, 13827–13832.
- Hedglin, M.; Aitha, M.; Pedley, A.; Benkovic, S.J. Replication protein A dynamically regulates monoubiquitination of proliferating cell nuclear antigen. J. Biol. Chem. 2019, 294, 5157–5168.
- Wu, X.; Yang, Z.; Liu, Y.; Zou, Y. Preferential localization of hyperphosphorylated replication protein A to double-strand break repair and checkpoint complexes upon DNA damage. Biochem. J. 2005, 391, 473–480.
- Ma, C.J.; Kwon, Y.; Sung, P.; Greene, E.C. Human RAD52 interactions with replication protein A and the RAD51 presynaptic complex. J. Biol. Chem. 2017, 292, 11702–11713.
- Stauffer, M.E.; Chazin, W.J. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J. Biol. Chem. 2004, 279, 25638–25645.
- Park, M.S.; Ludwig, D.L.; Stigger, E.; Lee, S.H. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 1996, 271, 18996–19000.
- Shinohara, A.; Shinohara, M.; Ohta, T.; Matsuda, S.; Ogawa, T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes. Cells 1998, 3, 145–156.
- Sugiyama, T.; Kowalczykowski, S.C. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002, 277, 31663–31672.
- Plate, I.; Hallwyl, S.C.; Shi, I.; Krejci, L.; Muller, C.; Albertsen, L.; Sung, P.; Mortensen, U.H. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J. Biol. Chem. 2008, 283, 29077–29085.
- Seong, C.; Sehorn, M.G.; Plate, I.; Shi, I.; Song, B.; Chi, P.; Mortensen, U.; Sung, P.; Krejci, L. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J. Biol. Chem. 2008, 283, 12166–12174.
- Liu, S.; Chu, J.; Yucer, N.; Leng, M.; Wang, S.Y.; Chen, B.P.; Hittelman, W.N.; Wang, Y. RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J. Biol. Chem. 2011, 286, 22314–22322.
- Gong, Z.; Chen, J. E3 ligase RFWD3 participates in replication checkpoint control. J. Biol. Chem. 2011, 286, 22308–22313.
- Nguyen, H.D.; Yadav, T.; Giri, S.; Saez, B.; Graubert, T.A.; Zou, L. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol. Cell 2017, 65, 832–847.e834.
- Yusufzai, T.; Kong, X.; Yokomori, K.; Kadonaga, J.T. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes. Dev. 2009, 23, 2400–2404.
- Ciccia, A.; Bredemeyer, A.L.; Sowa, M.E.; Terret, M.E.; Jallepalli, P.V.; Harper, J.W.; Elledge, S.J. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes. Dev. 2009, 23, 2415–2425.
- Yuan, J.; Ghosal, G.; Chen, J. The annealing helicase HARP protects stalled replication forks. Genes. Dev. 2009, 23, 2394–2399.
- Postow, L.; Woo, E.M.; Chait, B.T.; Funabiki, H. Identification of SMARCAL1 as a component of the DNA damage response. J. Biol. Chem. 2009, 284, 35951–35961.
- Kemp, M.G.; Akan, Z.; Yilmaz, S.; Grillo, M.; Smith-Roe, S.L.; Kang, T.H.; Cordeiro-Stone, M.; Kaufmann, W.K.; Abraham, R.T.; Sancar, A.; et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J. Biol. Chem. 2010, 285, 16562–16571.
- Unsal-Kacmaz, K.; Chastain, P.D.; Qu, P.P.; Minoo, P.; Cordeiro-Stone, M.; Sancar, A.; Kaufmann, W.K. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell Biol. 2007, 27, 3131–3142.
- Nagelhus, T.A.; Haug, T.; Singh, K.K.; Keshav, K.F.; Skorpen, F.; Otterlei, M.; Bharati, S.; Lindmo, T.; Benichou, S.; Benarous, R.; et al. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 1997, 272, 6561–6566.
- Machwe, A.; Lozada, E.; Wold, M.S.; Li, G.M.; Orren, D.K. Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. J. Biol. Chem. 2011, 286, 3497–3508.
- Hyun, M.; Park, S.; Kim, E.; Kim, D.H.; Lee, S.J.; Koo, H.S.; Seo, Y.S.; Ahn, B. Physical and functional interactions of Caenorhabditis elegans WRN-1 helicase with RPA-1. Biochemistry 2012, 51, 1336–1345.
- Brosh, R.M., Jr.; Orren, D.K.; Nehlin, J.O.; Ravn, P.H.; Kenny, M.K.; Machwe, A.; Bohr, V.A. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999, 274, 18341–18350.
- Shen, J.C.; Lao, Y.; Kamath-Loeb, A.; Wold, M.S.; Loeb, L.A. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech. Ageing Dev. 2003, 124, 921–930.
- Li, L.; Lu, X.; Peterson, C.A.; Legerski, R.J. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell Biol. 1995, 15, 5396–5402.
- Saijo, M.; Kuraoka, I.; Masutani, C.; Hanaoka, F.; Tanaka, K. Sequential binding of DNA repair proteins RPA and ERCC1 to XPA in vitro. Nucleic Acids Res. 1996, 24, 4719–4724.
- Matsuda, T.; Saijo, M.; Kuraoka, I.; Kobayashi, T.; Nakatsu, Y.; Nagai, A.; Enjoji, T.; Masutani, C.; Sugasawa, K.; Hanaoka, F.; et al. DNA repair protein XPA binds replication protein A (RPA). J. Biol. Chem. 1995, 270, 4152–4157.
- de Laat, W.L.; Appeldoorn, E.; Sugasawa, K.; Weterings, E.; Jaspers, N.G.; Hoeijmakers, J.H. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes. Dev. 1998, 12, 2598–2609.
- Matsunaga, T.; Park, C.H.; Bessho, T.; Mu, D.; Sancar, A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J. Biol. Chem. 1996, 271, 11047–11050.
- Bessho, T.; Sancar, A.; Thompson, L.H.; Thelen, M.P. Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J. Biol. Chem. 1997, 272, 3833–3837.
- Fisher, L.A.; Bessho, M.; Wakasugi, M.; Matsunaga, T.; Bessho, T. Role of interaction of XPF with RPA in nucleotide excision repair. J. Mol. Biol. 2011, 413, 337–346.
- Ellison, V.; Stillman, B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5’ recessed DNA. PLoS Biol. 2003, 1, E33.
- Guler, G.D.; Liu, H.; Vaithiyalingam, S.; Arnett, D.R.; Kremmer, E.; Chazin, W.J.; Fanning, E. Human DNA helicase B (HDHB) binds to replication protein A and facilitates cellular recovery from replication stress. J. Biol. Chem. 2012, 287, 6469–6481.
- Tkac, J.; Xu, G.; Adhikary, H.; Young, J.T.F.; Gallo, D.; Escribano-Diaz, C.; Krietsch, J.; Orthwein, A.; Munro, M.; Sol, W.; et al. HELB Is a Feedback Inhibitor of DNA End Resection. Mol. Cell 2016, 61, 405–418.
- Sparks, J.L.; Kumar, R.; Singh, M.; Wold, M.S.; Pandita, T.K.; Burgers, P.M. Human exonuclease 5 is a novel sliding exonuclease required for genome stability. J. Biol. Chem. 2012, 287, 42773–42783.
- Loo, Y.M.; Melendy, T. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J. Virol. 2004, 78, 1605–1615.
- Han, Y.; Loo, Y.M.; Militello, K.T.; Melendy, T. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 1999, 73, 4899–4907.
- Weisshart, K.; Taneja, P.; Fanning, E. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 1998, 72, 9771–9781.
- Arunkumar, A.I.; Klimovich, V.; Jiang, X.; Ott, R.D.; Mizoue, L.; Fanning, E.; Chazin, W.J. Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 2005, 12, 332–339.
- Park, C.J.; Lee, J.H.; Choi, B.S. Solution structure of the DNA-binding domain of RPA from Saccharomyces cerevisiae and its interaction with single-stranded DNA and SV40 T antigen. Nucleic Acids Res. 2005, 33, 4172–4181.
- Christensen, J.; Tattersall, P. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 2002, 76, 6518–6531.
- Yuzhakov, A.; Kelman, Z.; Hurwitz, J.; O’Donnell, M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999, 18, 6189–6199.
- Kim, H.S.; Brill, S.J. Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell Biol. 2001, 21, 3725–3737.
- Otterlei, M.; Warbrick, E.; Nagelhus, T.A.; Haug, T.; Slupphaug, G.; Akbari, M.; Aas, P.A.; Steinsbekk, K.; Bakke, O.; Krokan, H.E. Post-replicative base excision repair in replication foci. EMBO J. 1999, 18, 3834–3844.
- Loor, G.; Zhang, S.J.; Zhang, P.; Toomey, N.L.; Lee, M.Y. Identification of DNA replication and cell cycle proteins that interact with PCNA. Nucleic Acids Res. 1997, 25, 5041–5046.
- Dianov, G.L.; Jensen, B.R.; Kenny, M.K.; Bohr, V.A. Replication protein A stimulates proliferating cell nuclear antigen-dependent repair of abasic sites in DNA by human cell extracts. Biochemistry 1999, 38, 11021–11025.
- Farina, A.; Shin, J.H.; Kim, D.H.; Bermudez, V.P.; Kelman, Z.; Seo, Y.S.; Hurwitz, J. Studies with the human cohesin establishment factor, ChlR1. Association of ChlR1 with Ctf18-RFC and Fen1. J. Biol. Chem. 2008, 283, 20925–20936.
- Jeong, Y.T.; Rossi, M.; Cermak, L.; Saraf, A.; Florens, L.; Washburn, M.P.; Sung, P.; Schildkraut, C.L.; Pagano, M. FBH1 promotes DNA double-strand breakage and apoptosis in response to DNA replication stress. J. Cell Biol. 2013, 200, 141–149.
- Elia, A.E.; Wang, D.C.; Willis, N.A.; Boardman, A.P.; Hajdu, I.; Adeyemi, R.O.; Lowry, E.; Gygi, S.P.; Scully, R.; Elledge, S.J. RFWD3-Dependent Ubiquitination of RPA Regulates Repair at Stalled Replication Forks. Mol. Cell 2015, 60, 280–293.
- MacKay, C.; Toth, R.; Rouse, J. Biochemical characterisation of the SWI/SNF family member HLTF. Biochem. Biophys. Res. Commun. 2009, 390, 187–191.
- Sommers, J.A.; Banerjee, T.; Hinds, T.; Wan, B.; Wold, M.S.; Lei, M.; Brosh, R.M., Jr. Novel function of the Fanconi anemia group J or RECQ1 helicase to disrupt protein-DNA complexes in a replication protein A-stimulated manner. J. Biol. Chem. 2014, 289, 19928–19941.
- Cui, S.; Arosio, D.; Doherty, K.M.; Brosh, R.M., Jr.; Falaschi, A.; Vindigni, A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004, 32, 2158–2170.
- Cui, S.; Klima, R.; Ochem, A.; Arosio, D.; Falaschi, A.; Vindigni, A. Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J. Biol. Chem. 2003, 278, 1424–1432.
- Garcia, P.L.; Liu, Y.; Jiricny, J.; West, S.C.; Janscak, P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004, 23, 2882–2891.
- Hu, Y.; Raynard, S.; Sehorn, M.G.; Lu, X.; Bussen, W.; Zheng, L.; Stark, J.M.; Barnes, E.L.; Chi, P.; Janscak, P.; et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes. Dev. 2007, 21, 3073–3084.
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Jackson, S.P. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes. Dev. 2012, 26, 1179–1195.
- Dornreiter, I.; Erdile, L.F.; Gilbert, I.U.; von Winkler, D.; Kelly, T.J.; Fanning, E. Interaction of DNA polymerase Alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992, 11, 769–776.
- Feng, J.; Wakeman, T.; Yong, S.; Wu, X.; Kornbluth, S.; Wang, X.F. Protein phosphatase 2A-dependent dephosphorylation of replication protein A is required for the repair of DNA breaks induced by replication stress. Mol. Cell Biol. 2009, 29, 5696–5709.
- Majka, J.; Binz, S.K.; Wold, M.S.; Burgers, P.M. Replication protein A directs loading of the DNA damage checkpoint clamp to 5’-DNA junctions. J. Biol. Chem. 2006, 281, 27855–27861.
- Davis, A.P.; Symington, L.S. The Rad52-Rad59 complex interacts with Rad51 and replication protein A. DNA Repair 2003, 2, 1127–1134.
- Grimme, J.M.; Honda, M.; Wright, R.; Okuno, Y.; Rothenberg, E.; Mazin, A.V.; Ha, T.; Spies, M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic Acids Res. 2010, 38, 2917–2930.
- Bansbach, C.E.; Betous, R.; Lovejoy, C.A.; Glick, G.G.; Cortez, D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes. Dev. 2009, 23, 2405–2414.
- Yang, Z.G.; Liu, Y.; Mao, L.Y.; Zhang, J.T.; Zou, Y. Dimerization of human XPA and formation of XPA2-RPA protein complex. Biochemistry 2002, 41, 13012–13020.
- Stigger, E.; Drissi, R.; Lee, S.H. Functional analysis of human replication protein A in nucleotide excision repair. J. Biol. Chem. 1998, 273, 9337–9343.
- Daughdrill, G.W.; Buchko, G.W.; Botuyan, M.V.; Arrowsmith, C.; Wold, M.S.; Kennedy, M.A.; Lowry, D.F. Chemical shift changes provide evidence for overlapping single-stranded DNA- and XPA-binding sites on the 70 kDa subunit of human replication protein A. Nucleic Acids Res. 2003, 31, 4176–4183.
- Aboussekhra, A.; Biggerstaff, M.; Shivji, M.K.; Vilpo, J.A.; Moncollin, V.; Podust, V.N.; Protic, M.; Hubscher, U.; Egly, J.M.; Wood, R.D. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 1995, 80, 859–868.
- Wong, J.M.; Ionescu, D.; Ingles, C.J. Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. Oncogene 2003, 22, 28–33.
- Liu, J.; Doty, T.; Gibson, B.; Heyer, W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010, 17, 1260–1262.
- Yates, L.A.; Tannous, E.A.; Morgan, R.M.; Burgers, P.M.; Zhang, X. A DNA damage-induced phosphorylation circuit enhances Mec1(ATR) Ddc2(ATRIP) recruitment to Replication Protein A. Proc. Natl. Acad. Sci. USA 2023, 120, e2300150120.
- Din, S.; Brill, S.J.; Fairman, M.P.; Stillman, B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes. Dev. 1990, 4, 968–977.
- Binz, S.K.; Lao, Y.; Lowry, D.F.; Wold, M.S. The phosphorylation domain of the 32-kDa subunit of replication protein A (RPA) modulates RPA-DNA interactions. Evidence for an intersubunit interaction. J. Biol. Chem. 2003, 278, 35584–35591.
- Lee, S.; Heo, J.; Park, C.J. Determinants of replication protein A subunit interactions revealed using a phosphomimetic peptide. J. Biol. Chem. 2020, 295, 18449–18458.
- Stephan, H.; Concannon, C.; Kremmer, E.; Carty, M.P.; Nasheuer, H.P. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res. 2009, 37, 6028–6041.
- Zou, Y.; Liu, Y.; Wu, X.; Shell, S.M. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J. Cell Physiol. 2006, 208, 267–273.
- Byrne, B.M.; Oakley, G.G. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin. Cell Dev. Biol. 2019, 86, 112–120.
- Block, W.D.; Yu, Y.; Lees-Miller, S.P. Phosphatidyl inositol 3-kinase-like serine/threonine protein kinases (PIKKs) are required for DNA damage-induced phosphorylation of the 32 kDa subunit of replication protein A at threonine 21. Nucleic Acids Res. 2004, 32, 997–1005.
- Nuss, J.E.; Patrick, S.M.; Oakley, G.G.; Alter, G.M.; Robison, J.G.; Dixon, K.; Turchi, J.J. DNA damage induced hyperphosphorylation of replication protein A. 1. Identification of novel sites of phosphorylation in response to DNA damage. Biochemistry 2005, 44, 8428–8437.
- Patrick, S.M.; Oakley, G.G.; Dixon, K.; Turchi, J.J. DNA damage induced hyperphosphorylation of replication protein A. 2. Characterization of DNA binding activity, protein interactions, and activity in DNA replication and repair. Biochemistry 2005, 44, 8438–8448.
- Zernik-Kobak, M.; Vasunia, K.; Connelly, M.; Anderson, C.W.; Dixon, K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J. Biol. Chem. 1997, 272, 23896–23904.
More
