Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Catherine Yang and Version 2 by Catherine Yang.
Central to the development and survival of all organisms is the regulation of gene expression, which begins with the process of transcription catalyzed by RNA polymerases. During transcription of protein-coding genes, the general transcription factors (GTFs) work alongside RNA polymerase II (Pol II) to assemble the preinitiation complex at the transcription start site, open the promoter DNA, initiate synthesis of the nascent messenger RNA, transition to productive elongation, and ultimately terminate transcription.
- adenomyosis
- estrogen
- progesterone
- endocrine disruptors
- therapy
1. Introduction
Controlling gene expression is essential to normal growth, development, and sustained life. In metazoans, this requires regulating the spatial, temporal, and developmental expression of genes in a wide diversity of cell types. Mis-regulation of gene expression contributes to most disease states. The main control point for regulating gene expression is at the level of transcription. In eukaryotic cells, RNA polymerase II (Pol II) transcribes protein-coding genes into messenger RNA (mRNA) transcripts. Pol II also synthesizes long non-coding RNA (lncRNA) and most small nuclear RNA (snRNA) and microRNA (miRNA). Pol II transcription is vital for cell proliferation, proper expression of metabolic enzymes, signaling, cell fate, differentiation, gene expression, and nearly every cellular process. Although the RNA polymerase II transcription system is highly conserved across eukaryotes, this revisewarch is primarily focused on the human system, with some references to data from Drosophila and yeast systems.
The Pol II core enzyme can itself synthesize RNA using a template DNA, but promoter-specific transcription initiation requires the canonical general transcription factors (GTFs): TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (Table 1). In addition, the large multi-subunit complex Mediator is essential for proper transcription in cells [1][2]. Chromatin remodeling/modifying complexes and additional co-regulatory factors function together with promoter-specific transcriptional activators and repressors to set the proper level and timing of transcription from individual genes in specific cell types.
Table 1. Summary of the general transcription factors and RNA polymerase II.
| Protein | Subunits | Size (kDa) | Main Binding Partners | Function |
|---|---|---|---|---|
| TFIID | TBP, 13 TAFs | ~1300 | promoter, Pol II | Nucleates PIC assembly by binding multiple core promoter elements |
| TFIIA | TFIIAα, TFIIAβ, TFIIAɣ | 35, 19, and 12 | TBP, TFIID | Stabilizes the TFIID-DNA interaction; enhances the effects of transcriptional co-activators |
| TFIIB | TFIIB | 33 | promoter, TBP, Pol II | Helps to define the start site of transcription and orient Pol II in the proper direction |
| TFIIF | RAP30, RAP74 | 30 and 74 | promoter, Pol II, GTFs | Guides Pol II to the PIC and facilitates elongation |
| Pol II | Rpb1–Rpb12 | ~514 | promoter, all GTFs | Catalyzes RNA synthesis; phosphorylation of the CTD tail of Rpb1 serves a regulatory role |
| TFIIE | TFIIEα, TFIIEβ | 56 and 34 | promoter, TFIIH, Pol II, TFIIF | Recruits TFIIH to the PIC; stimulates enzymatic activities of TFIIH; stabilizes the open DNA conformation |
| TFIIH | Core domain: XPD, XBP, p62, p52, p44, p34, p8; CAK domain: CDK7, MAT1, cyclin H | ~500 | downstream DNA, TFIIE, Pol II | CDK7 kinase phosphorylates the CTD; ATP-dependent XPB translocase opens the promoter DNA |
2. RNA Polymerase II
Pol II is a large ~500 kDa complex made up of 12 protein subunits, named Rpb1-12. Studies have shown that 10 of the 12 subunits form the catalytic core of the Pol II complex and are either identical (Rbp5, 6, 8, 10, 12) or highly similar (Rpb1-3, 9, 11) to subunits found in RNA polymerase I and RNA polymerase III, which transcribe primarily tRNAs and rRNAs, respectively [3][4]. Recent ChIP-seq (chromatin immunoprecipitation followed by high-throughput sequencing) and mass spectrometry studies have shown that different sets of Rpb subunits differentially regulate select subsets of human genes, demonstrating the dense layers of regulation within the Pol II complex itself [5]. Crystal structures of yeast Pol II, and more recently cryo-EM structures of human and yeast Pol II, have revealed that the Pol II complex can be divided into the core, shelf, jaw lobe, and clamp structural domains that interact with each other and undergo conformational changes during the stages of transcription (these structures have been extensively reviewed in the literature [6][7][8][9]). The core domain contains Rpb3 and Rpb10-12 as well as the positively charged active center cleft, formed by Rpb1 and Rpb2 [10][11]. The active site of Pol II is buried deep at the base of the active center cleft, thus requiring translocation of the template DNA strand to the active site after entering the cleft. The shelf and jaw lobe elements have little observed movement but can rotate parallel to the active center cleft [11]. The clamp domain is connected to the active site cleft in the core domain through an array of flexible switches, and it swings nearly 30 Å upon opening or closing the cleft [3][11]. While not considered part of the catalytic core of Pol II, binding of the Rpb4 and Rpb7 subunits has been shown to be vital for maintaining the closed conformation of the Pol II clamp over the DNA during initiation [12][13]. It is hypothesized that the closing of this clamp domain over the cleft coupled with DNA distortion may facilitate promoter melting [14].
At 250 kDa, Rpb1 is the largest of all Pol II subunits and the principle catalytic subunit of the Pol II complex [15]. Beyond its catalytic role, Rbp1 plays a regulatory role in the transcription cycle that is mediated by the unstructured C-terminal domain (CTD) on the Rpb1 subunit. The CTD consists of a long tail comprising heptapeptide repeats of the consensus sequence YSPTSPS, with minor variability at the Ser7 position in repeats near the C-terminus [16][17]. Mammalian Pol II contains 52 repeats, with the number of repeats varying among different organisms in a manner that loosely correlates with genomic complexity [18]. The YSPTSPS consensus sequence is well conserved across eukaryotes, emphasizing the functional importance of each residue [18]. The CTD tail is not necessary for basal (i.e., unregulated) Pol II transcription in vitro [19][20]; however, it is required for accurate Pol II transcription and proper termination in cells [21][22][23]. The CTD is thought to function as a binding platform for association of numerous other protein complexes that help regulate co-transcriptional processes or steps in transcription, including RNA splicing and transcription termination [24][25][26].
The residues within the heptapeptide repeat are substrates for many post-translational modifications, with phosphorylation being the most well characterized. The Tyr, Ser, and Thr residues can be reversibly phosphorylated/dephosphorylated, allowing for regulation of Pol II activity through the transcription reaction and of Pol II CTD affinity for various regulatory factors [16][17][24] (Figure 1). For example, the level of specific phosphorylation marks varies across different stages of transcription, depending on the purpose of the modification. The use of phospho-specific antibodies coupled to ChIP-seq, in addition to in vitro work, have enhanced understanding of how CTD phosphorylation patterns change throughout the transcription cycle. Pol II is recruited to preinitiation complexes on promoter DNA in a hypo-phosphorylated form. Phosphorylation of Ser5 by the CDK7 kinase subunit of TFIIH (which is one of the general transcription factors discussed below) facilitates initiation of transcription. The Ser5 mark is removed as Pol II moves throughout the gene body. As a counterpoint, Ser2 phosphorylation predominantly accumulates after initiation to help recruit elongation and RNA processing factors and peaks at the 3’ ends of genes where it is thought to facilitate termination [27][28][29][30]. Beyond Ser5 and Ser2, other sites of phosphorylation on the Pol II CTD include Tyr1, Thr4, and Ser7, which have not been studied in the same detail as the other CTD residues. Research has shown that the significance of these three residues can vary across species, with metazoan and yeast systems sometimes exhibiting different behaviors [31][32]. Exploring the function of these other important Pol II CTD residues provides numerous areas for future study.
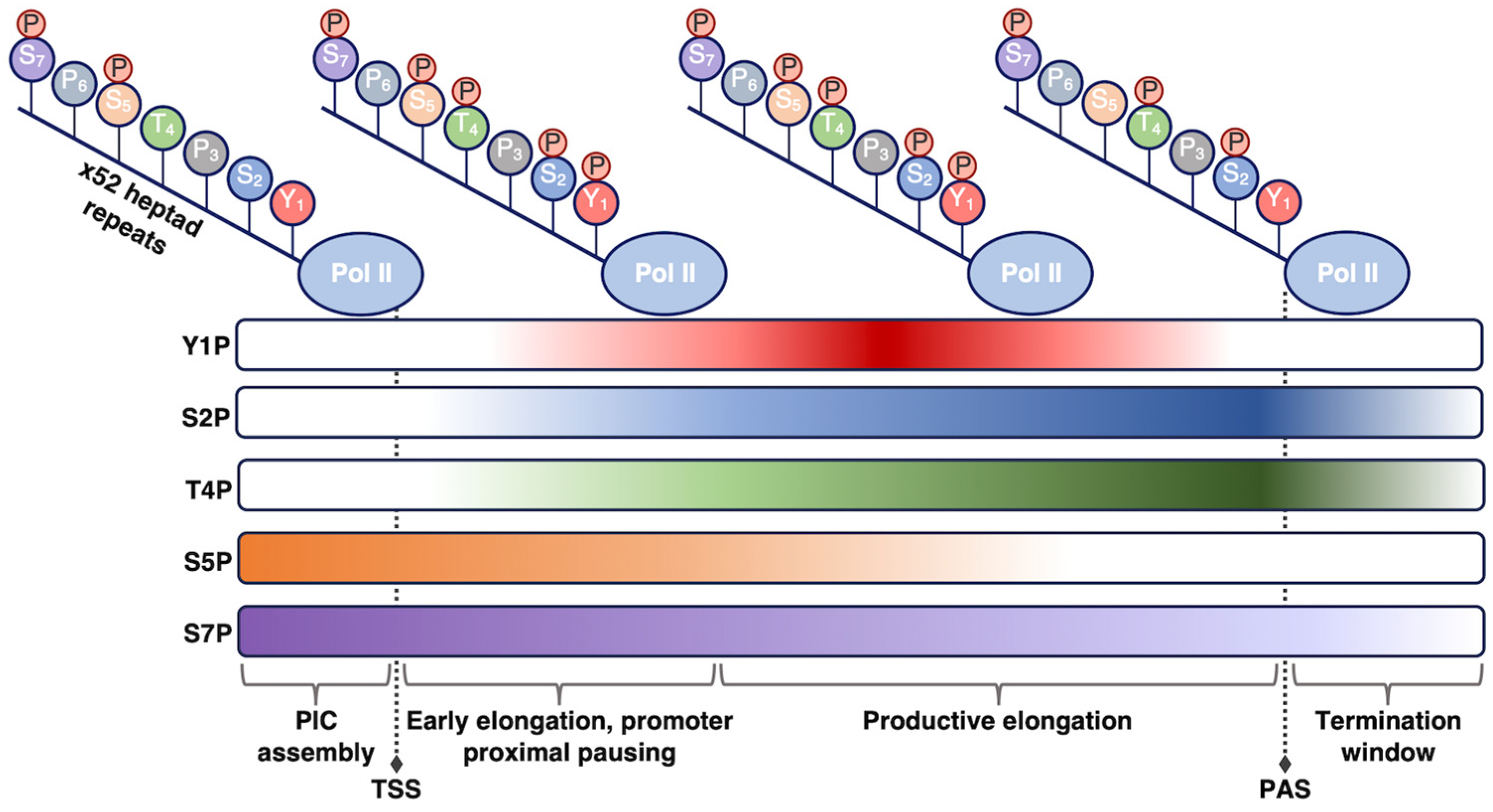
Figure 1. Phosphorylation state of the Pol II CTD is regulated during transcription. As Pol II transcribes through a gene and progresses through the stages of transcription (shown from left to right), different phosphorylation marks are added or removed to promote unique functions. The phosphorylation patterns shown here pertain to human Pol II; other organisms may exhibit slight differences in these patterns. TSS, transcription start site; PAS, polyadenylation site.
References
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The Mediator Complex as a Master Regulator of Transcription by RNA Polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 23, 732–749.
- Soutourina, J. Transcription Regulation by the Mediator Complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274.
- Cramer, P.; Armache, K.-J.; Baumli, S.; Benkert, S.; Brueckner, F.; Buchen, C.; Damsma, G.E.; Dengl, S.; Geiger, S.R.; Jasiak, A.J.; et al. Structure of Eukaryotic RNA Polymerases. Annu. Rev. Biophys. 2008, 37, 337–352.
- Vannini, A.; Cramer, P. Conservation between the RNA Polymerase I, II, and III Transcription Initiation Machineries. Mol. Cell 2012, 45, 439–446.
- Li, Y.; Huang, J.; Zhu, J.; Bao, L.; Wang, H.; Jiang, Y.; Tian, K.; Wang, R.; Zheng, H.; Duan, W.; et al. Targeted Protein Degradation Reveals RNA Pol II Heterogeneity and Functional Diversity. Mol. Cell 2022, 82, 3943–3959.e11.
- Osman, S.; Cramer, P. Structural Biology of RNA Polymerase II Transcription: 20 Years on. Annu. Rev. Cell Dev. Biol. 2020, 36, 1–34.
- Schier, A.C.; Taatjes, D.J. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Genes Dev. 2020, 34, 465–488.
- Aibara, S.; Schilbach, S.; Cramer, P. Structures of Mammalian RNA Polymerase II Pre-Initiation Complexes. Nature 2021, 594, 124–128.
- Schier, A.C.; Taatjes, D.J. Everything at Once: Cryo-EM Yields Remarkable Insights into Human RNA Polymerase II Transcription. Nat. Struct. Mol. Biol. 2021, 28, 540–543.
- Gnatt, A.L.; Cramer, P.; Fu, J.; Bushnell, D.A.; Kornberg, R.D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science 2001, 292, 1876–1882.
- Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nat. Struct. Mol. Biol. 2004, 11, 394–403.
- Armache, K.-J.; Kettenberger, H.; Cramer, P. Architecture of Initiation-Competent 12-Subunit RNA Polymerase II. Proc. Natl. Acad. Sci. USA 2003, 100, 6964–6968.
- Bushnell, D.A.; Westover, K.D.; Davis, R.E.; Kornberg, R.D. Structural Basis of Transcription: An RNA Polymerase II–TFIIB Cocrystal at 4.5 Angstroms. Science 2004, 303, 983–988.
- Dienemann, C.; Schwalb, B.; Schilbach, S.; Cramer, P. Promoter Distortion and Opening in the RNA Polymerase II Cleft. Mol. Cell 2019, 73, 97–106.e4.
- Bernecky, C.; Herzog, F.; Baumeister, W.; Plitzko, J.M.; Cramer, P. Structure of Transcribing Mammalian RNA Polymerase II. Nature 2016, 529, 551–554.
- Lyons, D.E.; McMahon, S.; Ott, M. A Combinatorial View of Old and New RNA Polymerase II Modifications. Transcription 2020, 11, 66–82.
- Harlen, K.M.; Churchman, L.S. The Code and beyond: Transcription Regulation by the RNA Polymerase II Carboxy-Terminal Domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273.
- Liu, P.; Kenney, J.M.; Stiller, J.W.; Greenleaf, A.L. Genetic Organization, Length Conservation, and Evolution of RNA Polymerase II Carboxyl-Terminal Domain. Mol. Biol. Evol. 2010, 27, 2628–2641.
- Zehring, W.A.; Lee, J.M.; Weeks, J.R.; Jokerst, R.S.; Greenleaf, A.L. The C-Terminal Repeat Domain of RNA Polymerase II Largest Subunit Is Essential in Vivo but Is Not Required for Accurate Transcription Initiation in Vitro. Proc. Natl. Acad. Sci. USA 1988, 85, 3698–3702.
- Kim, W.Y.; Dahmus, M.E. The Major Late Promoter of Adenovirus-2 Is Accurately Transcribed by RNA Polymerases IIO, IIA, and IIB. J. Biol. Chem. 1989, 264, 3169–3176.
- Gerber, H.-P.; Hagmann, M.; Seipel, K.; Georgiev, O.; West, M.A.L.; Litingtung, Y.; Schaffner, W.; Corden, J.L. RNA Polymerase II C-Terminal Domain Required for Enhancer-Driven Transcription. Nature 1995, 374, 660–662.
- Yahia, Y.; Pigeot, A.; El Aabidine, A.Z.; Shah, N.; Karasu, N.; Forné, I.; Krebs, S.; Blum, H.; Esnault, C.; Sexton, T.; et al. RNA Polymerase II CTD Is Dispensable for Transcription and Required for Termination in Human Cells. EMBO Rep. 2023, 24, e56150.
- Garg, A.; Sanchez, A.M.; Schwer, B.; Shuman, S. Transcriptional Profiling of Fission Yeast RNA Polymerase II CTD Mutants. RNA 2021, 27, 560–570.
- Singh, N.; Asalam, M.; Ansari, M.O.; Gerasimova, N.S.; Studitsky, V.M.; Akhtar, M.S. Transcription by RNA Polymerase II and the CTD-Chromatin Crosstalk. Biochem. Biophys. Res. Commun. 2022, 599, 81–86.
- Maita, H.; Nakagawa, S. What Is the Switch for Coupling Transcription and Splicing? RNA Polymerase II C-terminal Domain Phosphorylation, Phase Separation and Beyond. WIREs RNA 2020, 11, e1574.
- Jeronimo, C.; Bataille, A.R.; Robert, F. The Writers, Readers, and Functions of the RNA Polymerase II C-Terminal Domain Code. Chem. Rev. 2013, 113, 8491–8522.
- Davidson, L.; Muniz, L.; West, S. 3′ End Formation of Pre-mRNA and Phosphorylation of Ser2 on the RNA Polymerase II CTD Are Reciprocally Coupled in Human Cells. Genes Dev. 2014, 28, 342–356.
- Nojima, T.; Gomes, T.; Grosso, A.R.F.; Kimura, H.; Dye, M.J.; Dhir, S.; Carmo-Fonseca, M.; Proudfoot, N.J. Mammalian NET-Seq Reveals Genome-Wide Nascent Transcription Coupled to RNA Processing. Cell 2015, 161, 526–540.
- Ahn, S.H.; Kim, M.; Buratowski, S. Phosphorylation of Serine 2 within the RNA Polymerase II C-Terminal Domain Couples Transcription and 3′ End Processing. Mol. Cell 2004, 13, 67–76.
- Gu, B.; Eick, D.; Bensaude, O. CTD Serine-2 Plays a Critical Role in Splicing and Termination Factor Recruitment to RNA Polymerase II in Vivo. Nucleic Acids Res. 2013, 41, 1591–1603.
- Heidemann, M.; Eick, D. Tyrosine-1 and Threonine-4 Phosphorylation Marks Complete the RNA Polymerase II CTD Phospho-Code. RNA Biol. 2012, 9, 1144–1146.
- Yurko, N.M.; Manley, J.L. The RNA Polymerase II CTD “Orphan” Residues: Emerging Insights into the Functions of Tyr-1, Thr-4, and Ser-7. Transcription 2018, 9, 30–40.
More
