Replication protein A (RPA) is an ubiquitously expressed protein present in all eukaryotes
[1]. It plays essential roles in a wide range of DNA metabolic activities, including DNA replication, DNA recombination, DNA repair, and DNA damage signalling
[1][2][1,2]. RPA was initially identified in the Simian Virus 40 DNA replication system where it supported T-antigen-dependent DNA unwinding
[3][4][5][3,4,5]. RPA efficiently and with high affinity binds to single-stranded DNA (ssDNA) to avoid formations of secondary structures such as hairpins and to protect ssDNA against nuclease attacks
[1][6][7][8][9][1,6,7,8,9]. Thus, as the main eukaryotic ssDNA-binding protein, RPA safeguards ssDNA but also recruits various factors to the RPA–ssDNA complex such as ATRIP for ATR-dependent DNA damage signalling
[2][10][2,10]. Protein recruitment allows replication, checkpoint signalling, and repair pathways to occur
[10]. Post-translational modifications may lead to conformational changes in RPA, giving rise to additional functions and characteristics
[2][11][2,11].
2. RPA Structure, ssDNA and Protein Interactions
RPA is a heterotrimeric protein composed of subunits RPA70, RPA32, and RPA14 with apparent molecular weights of 70 kDa, 32 kDa, 14 kDa, respectively, as determined by SDS gel electrophoresis (
Figure 1 [1][5][12][1,5,12]). Each subunit contains at least one of the universally conserved oligonucleotide/oligosaccharide-binding (OB)-fold characterised by twisted β sheets and ⍺-helix capped ends. These OB-fold domains allow RPA to bind to ssDNA or target proteins involved in DNA metabolism and DNA damage signalling
[10][13][14][10,13,14]. The three subunits bind together through a heterotrimerisation core located in DNA-binding domain-C (DBD-C) of RPA70, DBD-D of RPA32, and RPA14 (DBD-E) facilitated by hydrophobic interactions of ⍺-helices in their OB-folds in a synergistic manner, as represented in
Figure 1A
[12][13][12,13] and shown in the RPA structures (
Figure 1B–E).
The DNA-binding domains (DBDs) A and B bind ssDNA with a groove composed by the loops L12 and L45 flanking the β strands β2 and β3 of the OB-fold (
Figure 1B;
[13]). These two binding sites each contain two highly conserved aromatic residues, that stack with DNA bases, and two conserved hydrophobic aa. When the linker regions between the DBDs are not occupied by DNA, they become susceptible to proteolysis
[13][15][13,16]. DBD-C contains crucial zinc-binding motifs for RPA structural stability, and its ssDNA-binding capability (
Figure 1B
[12][13][12,13]). RPA binds to ssDNA in three modes, an 8–10 nucleotide, 15–23 nucleotide, and 28–30 nucleotide mode
[13][14][13,14]. These modes are a mark for the flexibility of the RPA–ssDNA interactions and may allow the modulation of the RPA binding to ssDNA by external proteins such as RAD52 as previously discussed (
[2][14][16][17][2,14,17,18]; for further discussion.
It was previously shown that RPA preferentially binds to polypyrimidine sequences
[1][6][1,6]. The dissociation constant for poly(dT) was measured as K
D = ~1 nM, whereas sequences with a mixed base content and a length of 34 or 57 are bound with lower affinity, K
D = 40 nM and 10 nM, respectively
[1][6][8][9][1,6,8,9]. Recent studies using substrates with different oligo(dT) lengths and EMSA confirmed the modular binding approach suggesting that RPA binds dT
10 and dT
14 with micromolar affinities. RPA has an affinity of K
D = ~10 nM for dT
15 and dT
20, whereas substrates with 30 or more dTs are bound with affinities K
D = ~5 nM
[18][19]. Moreover, RPA has high affinities to natural occurring telomeric oligonucleotide sequences with a length of 18 nucleotides or longer K
D = ~1 nM whether they contain a G-quadruplex (G4)-forming sequence or not. In contrast, the CST (CTC1-STN1-TEN1) complex, an RPA-like protein complex with ssDNA-binding and telomere synthesis function (
[19][20][20,21].
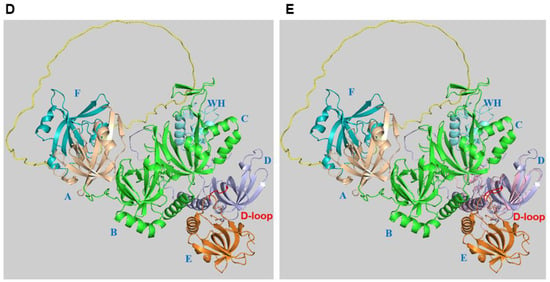
Figure 1. Structure of RPA. (
A) The bars represent the three RPA subunits with DNA-binding domains (DBDs) DBD-A to DBD-F highlighted and their borders indicated by amino acid numbering of the human protein sequences
[14][21][14,22]. Each DBD (colour code: RPA70: F (light brown), A (wheat), B and C (green); RPA32: D (light blue); RPA14: E (light orange)) contains a highly conserved OB-fold. The structure of one of the DBDs is highlighted in the bubble within the panel. The DBD domains A to D are able to bind to ssDNA. In contrast, the unstructured linker between DBD-F and A is shown in yellow, and the winged helix domain (WH) of RPA32 in light cyan are mainly protein–protein interaction sites. DBD-C contains the zinc-binding motifs represented by the protruding Zn. The N-terminus of RPA32, shown in dark blue and with the letter P, has recognition sites for multiple protein kinases including CDKs and PIKKs
[22][23]. The lines between the three subunits represent the points of interaction between the subunits through the heterotrimerisation core (adapted from
[12][23][12,24]). In the panel, the human RPA aa sequence is used for the numbering. (
B) The experimental RPA structure presents an image of the RPA heterotrimer including RPA70 (with DBD-A in wheat plus DBD-B and C in green, the associated Zn atom is shown in red), RPA32 in marine blue, RPA14 in light orange, and oligo(dT) ssDNA in orange with the 5′- and 3′-end marked (structure derived from PDB 4gop is in the 30 nt-binding mode
[13] and presented using Pymol (Schrödinger Inc. (USA)). In panel (
C), human RPA32 and STN1 OB-fold structures were predicted by AlphaFold
[24][25] and are shown in yellow light and magenta, respectively. STN1 is a subunit of the RPA-like CST complex and is involved in the initiation of G strand synthesis at telomeres
[19][20][20,21]. Both human structures were aligned to the experimental
Ustilago maydis RPA (structure derived from 4gop) using Pymol (for a better overview, aa N93 to T124 of STN1, which do not align to any structure, were omitted). The loops containing D157 of STN1, D151 of human RPA32, and D155 of
Ustilago maydis RPA32 are shown in red. Panels (
D,
E) present images of the RPA trimer predicted by AlphaFold
[24][25] without DNA and in a similar position as
Ustilago maydis RPA in panels (
B,
C). DBD-F, which is lacking in the
Ustilago maydis RPA structure, and DBD-A are in light green and light brown, respectively. The linker between the two domains is coloured in yellow. DBD-B, DBD-C and the short linker between the two domains are in green. The WH domain of RPA32, which is not shown in the
Ustilago maydis RPA structure, is presented in cyan. DBD-D plus the linker between DBD-D and WH are shown in light blue. The loop in DBD-D containing the conserved D151 is called the D loop (see also panels (
B,
C)) and is shown in red. It is important to note that the linker regions of RPA70 and RPA32 plus the N-terminal phosphorylation sequences of RPA32 have a very low per-residue model confidence score (pLDDT) and might be unstructured. To allow a better presentation of the RPA complex, the N-terminus of RPA32, aa 1–33, was omitted in panels (
D,
E). In panel (
E), the OB-fold of STN1 shown in light pink is aligned to human RPA using PyMol (for a better overview, aa N93 to T124 of STN1, which did not align to any structure, were omitted in the presentation). The loop containing the conserved D157 (D loop) is presented in red. The RPA domains are marked with blue letters, A to F (DBD-A to F) and WH in the structures.
To fulfil its functions, RPA not only binds to ssDNA but also to more than 50 proteins involved in DNA replication, repair, and recombination plus DNA damage signalling
[10][14][25][10,14,26]. Here, studies of the protein–protein binding sites of RPA have revealed that DBD-F and adjacent N-terminal sequences, which include the unstructured linker region and DBD-A, form a hub for protein interactions (
Figure 1A, and for their structural presentation, see
Figure 1D,
[10][14][21][25][26][10,14,22,26,27]). The N-terminal domain of RPA70, DBD-F, is involved in the binding of a variety of proteins in DNA replication, DNA damage response (DDR), and checkpoint signalling such as helicase B, WRN (Werner helicase), BLM (Bloom helicase), Tp53 (tumour-suppressor protein p53), RAD9 of RAD9–HUS1–RAD1 (9-1-1), ATR (ATM-Rad 3-related protein) ATRIP (ATR-interacting protein) and ETAA1, which is a newly identified ATR activator
[10][14][25][27][28][29][10,14,26,28,29,30]. Many of these contacts are mediated by the side pocket and the basic and hydrophobic groove of DBD-F
[10]. The adjacent linker region and DBD-A of RPA70 (see
Figure 1A,D) bind additional proteins such as RAD51, RAD52, Polα-p180, SV40 Tag (SV40 T antigen), and XPA
[14][25][30][31][14,26,31,32]. The WH domain of RPA32 is a second protein interaction hub of the RPA complex (see
Figure 1A,D) and binds to various proteins such as the DNA polymerase α (Pol α) subunit Prim2, RAD52, Tag, UNG protein, and XPA
[25][32][33][34][26,33,34,35]. Interestingly, the two protein binding hubs seem to be located on the same site of the RPA complex as predicted by AlphaFold. Additionally, the binding of RPA to human CDC45 and Primase-polymerase (hPrimpol1) regulates their cellular functions
[9][35][36][37][9,36,37,38].
3. Alternative RPA and Its Emerging Functions in Neurodegenerative Diseases
In addition to the subunits of ‘canonical’ heterotrimeric RPA, researchers have found in primates a RPA32-related protein, RPA4, a 30 kDa protein, which has 47% aa sequence identity and a 63% aa similarity with human RPA32 and can replace the latter in a complex with RPA70 and RPA14, forming a protein complex called alternative RPA (Alt-RPA)
[38][39][40][39,40,41]. RPA4 is mainly expressed in non-dividing cells and is preferentially detected in the bladder, esophagus, lung, placental, prostate, and colon mucosa tissue, but it is also found in human brain tissue
[38][39][39,40]. Interestingly, only primates express the RPA4 protein but not mice
[40][41][41,42]. Most biochemical activities of Alt-RPA are similar to those of canonical RPA. Alt-RPA stimulates SV40 Tag during the origin-dependent unwinding of dsDNA and DNA polymerase δ (Pol δ) as well as canonical RPA
[42][43][43,44]. However, Alt-RPA has weaker interactions with Pol α and is unable to stimulate DNA synthesis and de novo priming by Pol α during SV40 DNA replication in vitro
[42][43][43,44]. Additionally, Alt-RPA does not support cellular DNA replication
[39][40][43][40,41,44]. In contrast, Alt-RPA stimulates DNA synthesis by Pol α in the presence of RFC/PCNA, which is again contrary to canonical RPA.
4. Roles of RPA in DNA Replication
DNA replication is a process that requires the precise duplication of an organism’s genetic material once and only once in each cell division cycle
[44][45][49,50]. This process ensures that the daughter cells receive identical copies of the genetic material with each cell division, thereby maintaining genomic stability throughout numerous cell divisions
[45][50]. RPA plays a central and essential role in eukaryotic DNA replication through its ssDNA binding and protection plus protein recruitment functions
[44][49]. During the replication process, the MCM2-7 (minichromosome maintenance 2 to 7) complex is loaded onto origins of DNA replication with the help of ORC1-6 (origin recognition complex 1 to 6), Cdc6 (cell division cycle 6), and Cdt1. The activation of the MCM2-7 requires the loading of Cdc45 plus the GINS complex to chromatin and the formation of the CMG complex, the eukaryotic replicative helicase, which is supported by the proteins DONSON and DNA polymerase ε (Pol ε)
[46][47][48][49][50][51][52][53][51,52,53,54,55,56,57,58]. At the beginning of the S phase, CMG helicase unwinds dsDNA at the origins, and the dsDNA is separated into two ssDNA templates to allow the DNA polymerase to have access to the genetic information and to allow the duplication of DNA (summarised in
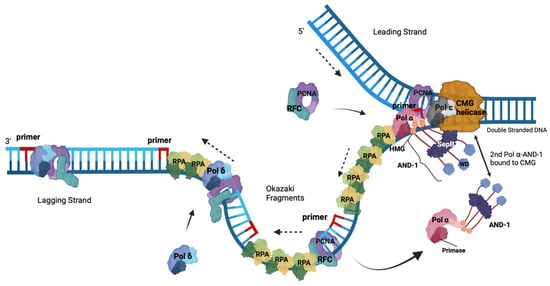
Figure 2 [44][49]). With the help of Cdc45, RPA binds to the exposed ssDNA in a polar fashion and stabilises the ssDNA
[9][36][44][54][9,37,49,59]. Due to the polar orientation of the DNA, at replication forks, the unwound DNA strands give rise to template DNAs with a 5′–3′ and a 3′–5′ direction. Together with the knowledge that DNA polymerases only synthesize DNA in 5′–3′ direction with the template having a 3′–5′ directionality, the DNA is synthesised continuously, forming the leading strand, whereas the second strand is synthesised discontinuously in small pieces called Okazaki fragments and forms the lagging strand as presented in
Figure 2 [44][49].
Figure 2. DNA synthesis at a eukaryotic replication fork. This schematic drawing of a eukaryotic replication fork shows CMG helicase (orange) unwinding dsDNA into the leading plus lagging strand templates and the replication proteins involved in DNA synthesis at the fork. Heterotrimeric RPA binds the resulting ssDNAs. The navy blue DNA strands are the parental strands. Pol ε (grey) newly synthesises the leading strand (royal blue DNA) in the 5′ to 3′ direction as indicated by the dashed arrow, and it is associated with the CMG. In contrast, the homotrimeric AND-1-CTF4-WDHD1 protein complex (dark grey, named AND-1) links CMG to the Pol α complex. The primer (red) synthesised by the primase function of Pol α allows for replication synthesis initiation by the DNA polymerase activity of Pol ⍺ (pink) in a 5′–3′ direction to start Okazaki fragment synthesis for lagging strand synthesis. PCNA (purple) and RFC (turquoise) replace Pol ⍺, making a landing site for Pol δ (maroon) to bind to the RNA–DNA and PCNA and elongate it until this complex reaches the next Okazaki on the parent strand, allowing the maturation of the Okazaki fragments to occur (adapted from
[44][47][55][56][49,52,60,61]). The solid arrows represent incoming and leaving proteins.
No replicative eukaryotic DNA polymerase starts DNA synthesis de novo. Therefore, a special enzyme, primase (often also called DNA primase), an RNA polymerase, synthesises short oligoribonucleotides with a length of ~10 nts or multimers thereof on ssDNA templates including unwound origin DNA to start DNA synthesis. Primase is an essential enzyme for DNA replication and functions as a dimer consisting of PRI1/Prim1/p49, the catalytic subunit, and PRI2/Prim2/p58, the regulatory subunit, which controls primer lengths
[57][58][59][62,63,64]. The primase dimer forms a heterotetrameric complex with the two larger subunits PolA1/p180 carrying the catalytic DNA polymerase activity and PolA2/p68/p70/B subunit, the second regulatory subunit, which is called DNA polymerase ⍺ (Pol ⍺) or DNA polymerase ⍺-primase
[60][65]. Pol α is the only replicative DNA polymerase complex having primase activity and thus is central for DNA replication processes. Under special circumstances, such as replication fork restart, Primpol with the help of RPA may take over the restart function
[35][61][36,66]. However, Pol α in association with the CST complex may also function in the replication restart
[62][67]. Primer synthesis is initiated by the cooperation of CMG, AND-1-Ctf4-WDHD1, RPA, Pol ⍺ and primase at 5′ ssDNA (
Figure 2 [47][48][63][52,53,68]).
During lagging strand synthesis, each Okazaki fragment is initiated by Pol α as described above. Taking the Okazaki fragment size and the human genome size into account, Pol α synthesises approximately 20 to 40 million primers to produce a complete round of lagging strand synthesis per cell division cycle, which is an enormous task
[64][75]. In the following, the RNA primer is handed over from the primase to PolA1/p180 by PRI2/Prim2 with the support of RPA, whereas the RNA–DNA primer is then transferred from Pol α to Pol δ
[60][65][66][65,70,72]. The latter synthesises the Okazaki fragment until it reaches the RNA moiety of the previous Okazaki fragment. Importantly, RPA needs to protect exposed ssDNA until pol δ encounters the next primer, and a DNA ligase ligates the fragments together
[56][66][61,72]. After reaching the previous Okazaki fragment, Pol δ continues the synthesis of the associated Okazaki fragment into the previous fragment in a strand displacement mode. Thus, by removing the RNA primer and a part of the Pol α-synthesised DNA, the enzymes create a 5′-flap DNA, which for efficient ligation of the two Okazaki fragments needs to be cleaved off
[56][66][61,72]. The enzyme flap endonuclease 1 (FEN1) takes over this role. FEN1 binds to the 5′-flap DNA and cleaves the DNA precisely at the ssDNA–dsDNA transition, leaving a nicked dsDNA remaining, which is an ideal substrate for LIG1 ligation
[56][66][61,72].
5. The Role of the RPA-Related, ssDNA-Binding Protein Complex CST in the Initiation of Okazaki Fragment Synthesis
To understand the initiation of Okazaki fragment synthesis and to elucidate the role of RPA in this process, it is important to know that primase has lower affinities for ssDNA than RPA, and it has been shown that RPA efficiently inhibits the primase activity of Pol α on natural ssDNA templates
[67][68][69][70][71][77,78,79,80,81]. In the SV40 DNA replication, a mechanism for the reversion of the inhibition has been described. Similar to the interaction of RAD52 with the RPA32 WH domain, Tag interacts with the WH domain, reverses the RPA inhibition, and allows primase to synthesise primers on RPA-bound ssDNA
[68][72][73][78,82,83]. One of the main functions in this process is the remodelling of the Pol α–primase complex from an inactive state to a priming active state
[20][64][74][75][76][21,75,84,85,86]. This process is best understood in the initiation process of Okazaki fragment synthesis on the G strand of telomeres as part of the C strand synthesis
[20][64][74][75][76][21,75,84,85,86]. The structure of Pol α in association with the CST complex, an RPA-like complex also called alpha-accessory factor (AAF)
[77][87], shows that the two protein complexes have numerous interactions with each other and some might be important for the remodelling of the Pol α complex to a pre-initiation complex
[20][64][21,75].
Recently, it was shown that the STN1 subunit of CST and RPA32 alone are sufficient to stimulate Pol α initiation activity and increase its primase activity on ssDNA
[67][78][77,88]. The OB-fold domain of human STN1 is sufficient to provide the stimulation of Pol α, and here, the conserved aa D157 of STN1, which is equivalent to D151 in human RPA32, is important for the stimulatory activity, whereas the ssDNA-binding activity of STN1 is not important
[67][77]. In the published pre-initiation complex of CST and Pol α, D157 interacts with the C-terminus of PolA1 (S1365 and R1366) and might be important for the remodelling of Pol α from an inactive to an active form and thus provide the stimulatory function for Pol α during the primase reaction
[64][67][74][75,77,84].
6. Functions of RPA in DNA Damage Response Pathways
DNA replication is a complex process, and errors commonly occur despite the proof-reading exonuclease activity associated with the replicative polymerases Pol δ and ε
[60][65]. Additionally, the genetic information of cells is under constant pressure from intracellular and extracellular/environmental stresses causing DNA damage. To preserve genomic stability, eukaryotic cells have developed multiple DNA repair pathways depending on the type of DNA damage to correct damaged DNA
[60][65]. Moreover, conserved DNA damage signalling pathways are present in eukaryotic cells to maintain genome stability. All these processes require RPA to fully function
[14][16][21][60][79][14,17,22,65,90].
-
Base excision repair (BER) involves the repair of damaged or modified bases in the DNA
[60][79][65,90].
-
Nucleotide excision repair (NER) is required for the removal of bulky DNA lesions induced, e.g., by UV radiation or chemicals
[60][79][65,90].
-
Mismatch repair (MMR) is responsible for the identification and elimination of mispaired nucleotides after DNA replication
[60][79][65,90].
-
Double-strand breaks (DSBs) repair has evolved in eukaryotic cells as two main pathways, homologous recombination (HR) and non-homologous end joining (NHEJ). HR utilizes a homologous DNA template to allow for the accurate repair of DSBs whereas NHEJ only re-joins the broken ends, opening the possibility of the induction of insertions or deletions
[60][79][65,90].
6.1. RPA in DNA Damage Signalling
Since DNA lesions are detrimental to eukaryotic cells and the stability of their genetic information, a fast and precise response to damaged DNA is required to correct DNA lesions in the chromosomal DNA of cells to avoid the appearance of genetic diseases such as cancer [2][10][2,10].
During DNA damage, the three RPA subunits are ubiquitinated. Specifically, RPA32 is modified by K63-linked ubiquitin chains which are important for ATRIP recruitment and ATR kinase activation [2]. DOCK7 is one protein that is phosphorylated by this ATR activation. In turn, Dock7 phosphorylation increases RPA association to chromatin, which again allows further ATR activation, essentially creating a positive feedback loop [80][91]. Cyclin-dependent kinases (CDKs) and phosphatidylinositol-3 kinase-related kinases (PIKKs) such as ATR, ATM (ataxia telangiectasia-mutated), and DNA-PK (DNA-dependent protein kinase) phosphorylate serine and threonine residues in the RPA32 N-terminal region during the cell cycle and in response to genotoxic stress [2][26][81][82][2,27,94,95]. There are eight phosphorylation sites in this RPA32 sequence where CDK phosphorylates S23 and S29, which then stimulates the ATR-dependent phosphorylation of S33.
6.2. RPA in DNA Repair Pathways
To avoid genome instability by bulky DNA adducts or major DNA structure distortions, nucleotide excision repair (NER) is an important DNA repair pathway (summarised in
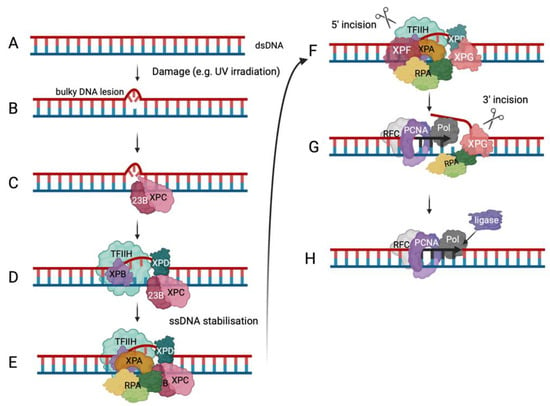
Figure 3 [83][84][85][97,98,99]). The importance of the NER pathway is underlined by the existence of a rare genetic disorder called Xeroderma pigmentosum (XP), in which genes coding for proteins involved in the NER pathway are mutated
[84][98]. These proteins are called XPA to XPG and XPV, which is a DNA polymerase called Pol η
[84][85][98,99].
Figure 3. Nucleotide excision repair. The schematic drawing depicts the global genomic NER (ggNER) pathway as the most prominent NER pathway in cells
[83][84][85][97,98,99]. Eukaryotic cells also repair bulky lesions via the transcription-coupled NER (tcNER) pathway, which only occurs in a transcription-dependent manner on the transcribed strand
[83][84][85][97,98,99]. The tcNER pathway feeds into the presented pathway but was omitted for simplification reasons. (
A) DNA damage can result in bulky DNA lesions. (
B) RAD23B and XPC (pink) recognise these damaged sites in dsDNA. (
C) They recruit the multi-subunit protein complex TFIIH (transcription facto II H, light green) that contains XPB in purple and XPD in dark green to verify and extend the opening of the dsDNA. (
D) XPA, in orange, and the multi-coloured RPA are recruited to the DNA damage site to bind and protect newly unwound ssDNA. The positioning of RPA and XPA ensures the correct localisation of XPF and XPG, in darker pink colours, to undertake their incision functions as shown in the panel. (
E–
G) DNA elongation occurs after binding of RFC (grey) and loading of PCNA (purple) and DNA polymerase (dark grey) recruitment (panel
G). (
H) Ligase (purple) binds the newly synthesised DNA at the incision site and ligates the nick in the dsDNA to yield repaired dsDNA (adapted from Schärer, 2013
[85][99]).
7. Conclusions
The central roles of RPA in a variety of metabolic processes show the importance of future research using different angles regarding RPA functions and cooperation with replication, repair and recombination proteins. The main f prospects will be the examination of RPA levels and/or post-translational modification as biomarkers and testing its inhibition to combat cancers and other diseases. Additionally, the structural biological insights into the various metabolic processes including RPA at replication forks and RPA in complex with DNA recombination mediators on ssDNA, to name a few, will significantly enhance our understanding of the roles of RPA and such can be harnessed for drug development. enhance the field and give insights into the mechanisms of these central reactions.