Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Jessie Wu and Version 2 by Jessie Wu.
The rhizosphere, the narrow zone of soil influenced by the plant root system, is a dynamic environment where complex interplay between plants and soil microbes occurs, and it may contain up to 1011 cells/g of root, with more than 30,000 bacterial species. There are various signals in the rhizosphere, including QS signals among microorganisms and root exudate signals from plants to microorganisms.
- rhizobacteria
- root exudation
- root colonization
- root exudate–rhizobacteria interaction
1. Introduction
Numerous studies have shown that root exudates can attract beneficial microorganisms, shape the rhizosphere microbiome, and improve plant performance [1][2]. In general, plants release 11–40% of their photosynthetic yield into the rhizosphere in the form of root exudates [3], which can be categorized as low- or high-molecular-weight compounds. Low-molecular-weight compounds encompass diverse molecules, such as sugars, organic acids, amino acids, alcohols, volatile compounds, and other secondary metabolites. High-molecular-weight compounds, such as mucilage (polysaccharides) and proteins, may exhibit less variability but usually constitute a larger fraction of exudates [4]. Most low-molecular-weight compounds, such as sugars, amino acids, and ions, are passively released via diffusion, channel, and vesicle transport. For example, the efflux of sugars is facilitated by “sugars will eventually be exported transporters” (SWEETs) along a concentration gradient [5][6]. However, other compounds, such as secondary metabolites, proteins, and polysaccharides, are generally secreted into the rhizosphere by an active mechanism via different membrane-bound proteins. For example, secondary metabolites, such as flavonoids, phenolics, and hormones, are actively secreted into the rhizosphere with the help of ATP-binding cassette (ABC) transporters or multidrug and toxic compound extrusion (MATE) transporters [7][8]. Notably, citrate, a low-molecular-weight compound, is also transported by MATE antiporters [9].
2. Effects of Plant Genotype and Development on Root Exudate–Rhizobacteria Interactions
The composition and quantity of root exudates are not static or uniform and change dynamically based on several factors. First, root exudates are determined by plant genotype (Figure 21A). As the distance between microorganisms and roots decreases (from rhizosphere soil to the endorhizosphere), the influence of plant genotype on the enrichment of microorganisms increases [10]. Many metabolites are shared across different species, while others are specific to certain plant species. For instance, benzoxazinoids (BXs), a class of defensive secondary metabolites, are predominantly released by the crown roots of different grass species, such as maize and wheat [1]. In addition, rhizobacteria exhibit diverse responses to root exudates from various plant genotypes [11]. This phenotypic trait allows the use of an intercropping system to improve the rhizosphere environment and establish a healthier microbial community. A recent study on an intercropping system involving tomato and potato onion revealed distinct exudation patterns for each species. The presence of raxifolin in potato onion root exudates alters the chemistry of tomato root exudates, leading to the recruitment of beneficial soil bacteria, such as Bacillus sp., and the establishment of a healthy rhizosphere [12] (Figure 1A).
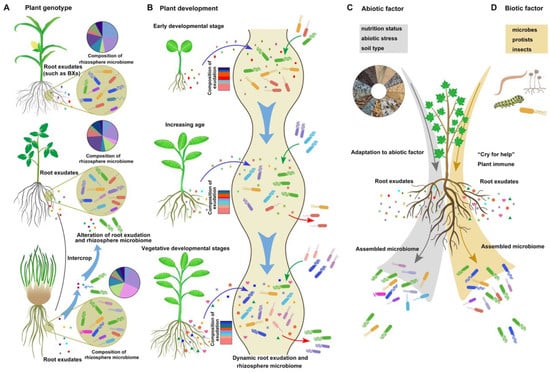
Figure 1. The influencing factors on root exudation and rhizosphere bacteria composition. The root exudation–rhizobacteria interaction is a dynamic process impacted by plant genotype (A), plant development (B), environmental abiotic factors (C), and biotic factors (D): (A) The process of root exudation displays variability across different plant genotypes. The diverse signaling receptors of bacteria and their ability to utilize and decrypt substances contribute to changes in the rhizosphere microbiome in the rhizosphere with different compositions of root exudates. (B) The blue arrow represents dynamic root exudation at different plant developmental stages. The green and red arrows indicate the attraction and repulsion of rhizobacteria, respectively. (C,D) In response to abiotic or biotic stress, plants can recruit some beneficial bacteria through exudation.
Furthermore, the composition of root exudates is influenced by plant diversity. High plant biodiversity is typically associated with high microbial diversity due to diverse exudates [7]. Essarioui et al. [13][14] reported that Streptomyces isolates, which are well known for producing antibiotics, from soil with polyculture plant species have larger niche widths than those from monoculture soil. In contrast, pathogenic Fusarium exhibits a greater niche preference in monocultures than in polycultures. The differences in the utilization of diverse exudates by beneficial or pathogenic bacteria may contribute to this phenomenon. Leveraging this phenomenon, researchers can potentially manipulate the soil microbial community by adopting different cropping patterns.
Second, root exudates are dynamic and change according to plant development (Figure 1B). A recent study demonstrated that Avena barbata produced higher levels of sucrose and homoserine during the early developmental stage than during other stages. With increasing age, the exudation of amino acids and carboxylic acids increases. At the vegetative development stage, A. barbata exhibited the greatest overall root exudation. During plant senescence, the abundance of quaternary ammonium salts and hormones significantly increases [3]. Similarly, in Arabidopsis, the highest abundance of sugars occurs at the early stage, predominantly used for root growth maintenance, while amino acids and phenolic exudation increase at the late plant developmental stage [15][16]. The rhizosphere microbial community and function are influenced by altered root exudates throughout different plant developmental stages [15].
Plants adjust their exudation patterns at different developmental stages to regulate the rhizosphere microbiome and fulfill the nutrient requirements of different growth periods [16]. Moreover, different bacterial communities are recruited by plants during various developmental stages, reflecting distinct choices in root exudate utilization (Figure 1B). Different plant species and development can selectively enrich specific microbial taxa that have the right transporters and enzymes to metabolize those specific root exudates, thereby shaping the microbiome composition. For instance, isolates recruited by A. barbata during the root growth stage display a preference for sugars, amino acids, organic acids, and quaternary amines [3]. Consequently, the nutrient preferences of bacterial strains lead to the enrichment of specific microbiome compositions at different stages of plant development.
3. Effects of Environmental Abiotic and Biotic Factors on Root Exudate–Rhizobacteria Interactions
Third, exudation is modulated by various environmental abiotic factors, including nutrient status, soil type, and abiotic stressors (Figure 1C). Nutrient deficiencies or excesses can alter the composition and quality of root exudates, potentially impacting the selection and growth of rhizobacterial communities. For example, coumarins, which are always induced by iron starvation, are phenolic secondary metabolites produced in roots and exuded into the rhizosphere in a MYB72/BGLU42-dependent manner in Arabidopsis [17]. Coumarins are necessary for beneficial interactions between Arabidopsis plants and the root microbiota under iron starvation conditions and can modulate the root microbial community, elicit microbe-associated iron nutrition, improve plant iron nutrition, and aid in the health of the soil rhizosphere [18]. The availability and composition of nutrients in the rhizosphere are also determined by the soil structure, such as the soil matrix and soil aggregates [19]. A rhizosphere with high nutrient availability may increase microbial carbon metabolism and facilitate the development of complex networks of soil rhizosphere microorganisms [20]. Soil pH, an important characteristic of soil type, plays a crucial role in determining the availability and effectiveness of root exudates, as well as the survival and growth of rhizobacteria [21][22]. Wan et al. [21] reported that soil pH has a greater influence on rhizobacterial communities than other physicochemical variables and vegetation types. Guo et al. [23] suggested that the soil microbial network in the rice rhizosphere becomes more stable and complex at higher pH levels (pH > 7.5), which is associated with improved efficiency of nutrient cycling efficiency. Additionally, plants can modulate soil pH by regulating the composition of root exudates, thereby affecting the presence of beneficial and pathogenic bacteria [22].
Various abiotic stressors, such as salinity, heavy metals, pollutants, and drought, substantially influence root exudate–rhizobacteria interactions. These stressors can modify root exudation patterns, impair the viability of rhizobacteria, and disrupt the symbiotic relationship between plants and rhizobacteria [24]. For instance, salt stress can lead to the dominance of certain organic acids as major components in the root exudates of the halophyte Limonium sinense, promoting the growth and chemotaxis of the beneficial strain B. flexus KLBMP 4941 and subsequently benefiting plant growth under salt stress [25]. Under heavy metal stress, more exudates, particularly organic nitrogen, are released to attract the beneficial bacterium P. putida E36, which aids Miscanthus × giganteus plants in coping with heavy metal stress [26]. The abundance of organic acids and amino acids in rice exudates increased in response to chlorpyrifos stress, attracting the beneficial soil bacterium Sphingomonas sp. to degrade chlorpyrifos [27]. In addition, soil moisture levels are the key factors affecting the quantity and quality of root exudation and rhizobacterial colonization [28]. For instance, drought stress leads to increased production of many metabolites secreted by roots, with a notable increase in glycerol-3-phosphate. Altered plant metabolites enhance the abundance and activity of monoderm bacteria, which in turn improve plant fitness under drought stress [29]. By considering and managing these abiotic factors, researchers can enhance the efficiency and effectiveness of rhizobacterial colonization and maximize the benefits derived from this symbiotic relationship.
Fourth, exudation is influenced by various environmental biotic factors. Specific changes in root exudation patterns can be induced by different microbes, protists, and insects, such as many secondary metabolites and small peptides (Figure 1D). Differential immune and metabolic responses occur in roots when these plants interact with beneficial and pathogenic microbes, leading to either the promotion or suppression of colonization. A recent study showed that glycosylated azelaic acid, which is induced by the soil microbiome, may act as a potential microbe-induced signal in reprogramming systemic root exudates [30]. Some altered compounds in root exudates exhibit variable antimicrobial activity and can function as plant bioprotectants against pathogens and as attractants for certain plant-associated bacteria [4]. Increasing evidence indicates that upon colonization by beneficial bacteria, plants activate specific immune responses, while certain beneficial bacteria exhibit mechanisms to tolerate root immune responses through spatial evasion and high tolerance strategies [2]. The immune response activated by the beneficial strain Pseudomonas varies at different sites in the root, suggesting that the spatial mitigation of the colonization site may contribute to Pseudomonas sp. evasion of root immunity [31][32]. The PGPR B. velezensis SQR9 can tolerate the bursts of ROS through the specific two-component regulatory system ResDE [33]. Additionally, upon colonization by beneficial bacteria, the presence of many compounds exuded into root exudates, such as phenolics, acyl sugars, and some antimicrobial compounds, changes to improve beneficial bacterial colonization and inhibit pathogen colonization [30][34][35].
After pathogen infection or herbivore damage, plants can “cry for help” with root exudates and regulate plant immune signaling. The altered composition of root exudates caused by pathogens or herbivores leads to an increase in antimicrobial substances specific to the pathogen, as well as chemoattractants that attract beneficial rhizobacteria. For instance, upon infection by the fungal pathogen Fusarium culmorum, the blend of VOCs emitted by Carex arenaria roots changes, and some specific bacteria with antifungal properties are attracted. Among the altered VOCs, several substances, such as benzofuran and acetophone, exhibit antifungal activity [36]. Furthermore, the infection of ginger by the pathogen Ralstonia solanacearum enhances the secretion of several antibacterial compounds by gingers and increases the abundance of beneficial and stress-tolerant bacteria, ultimately inhibiting ginger bacterial wilt [37]. In potato plants, aphid infection reduces the glucose and fructose levels in root exudates [38]. In addition, the phytohormone salicylic acid not only induces plant immune signaling in response to pathogens but also modulates colonization by specific rhizobacteria [39]. These findings support the notion that some defense signals activated by pathogen infection might promote some beneficial bacterial colonization.
References
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root Exudate Metabolites Drive Plant-Soil Feedbacks on Growth and Defense by Shaping the Rhizosphere Microbiota. Nat. Commun. 2018, 9, 2738.
- Liu, Y.; Xu, Z.; Chen, L.; Xun, W.; Shu, X.; Chen, Y.; Sun, X.; Wang, Z.; Ren, Y.; Shen, Q.; et al. Root Colonization by Beneficial Rhizobacteria. FEMS Microbiol. Rev. 2024, 48, fuad066.
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; Da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic Root Exudate Chemistry and Microbial Substrate Preferences Drive Patterns in Rhizosphere Microbial Community Assembly. Nat. Microbiol. 2018, 3, 470–480.
- Chagas, F.O.; Pessotti, R.D.C.; Caraballo-Rodríguez, A.M.; Pupo, M.T. Chemical Signaling Involved in Plant-Microbe Interactions. Chem. Soc. Rev. 2018, 47, 1652–1704.
- Breia, R.; Conde, A.; Badim, H.; Fortes, A.M.; Gerós, H.; Granell, A. Plant Sweets: From Sugar Transport to Plant–Pathogen Interaction and More Unexpected Physiological Roles. Plant Physiol. 2021, 186, 836–852.
- Lin, I.W.; Sosso, D.; Chen, L.-Q.; Gase, K.; Kim, S.-G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.-H.; Qu, X.-Q.; et al. Nectar Secretion Requires Sucrose Phosphate Synthases and the Sugar Transporter SWEET9. Nature 2014, 508, 546–549.
- Sasse, J.; Martinoia, E.; Northen, T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018, 23, 25–41.
- Liu, L.; Zhao, L.; Chen, P.; Cai, H.; Hou, Z.; Jin, X.; Aslam, M.; Chai, M.; Lai, L.; He, Q.; et al. ATP Binding Cassette Transporters ABCG1 and ABCG16 Affect Reproductive Development via Auxin Signalling in Arabidopsis. Plant J. 2020, 102, 1172–1186.
- Furukawa, J.; Yamaji, N.; Wang, H.; Mitani, N.; Murata, Y.; Sato, K.; Katsuhara, M.; Takeda, K.; Ma, J.F. An Aluminum-Activated Citrate Transporter in Barley. Plant Cell Physiol. 2007, 48, 1081–1091.
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424.
- Haney, C.H.; Samuel, B.S.; Bush, J.; Ausubel, F.M. Associations with Rhizosphere Bacteria Can Confer an Adaptive Advantage to Plants. Nat. Plants 2015, 1, 15051.
- Zhou, X.; Zhang, J.; Khashi u Rahman, M.; Gao, D.; Wei, Z.; Wu, F.; Dini-Andreote, F. Interspecific Plant Interaction via Root Exudates Structures the Disease Suppressiveness of Rhizosphere Microbiomes. Mol. Plant 2023, 16, 849–864.
- Essarioui, A.; Kistler, H.C.; Kinkel, L.L. Nutrient Use Preferences among Soil Streptomyces Suggest Greater Resource Competition in Monoculture than Polyculture Plant Communities. Plant Soil 2016, 409, 329–343.
- Essarioui, A.; LeBlanc, N.; Kistler, H.C.; Kinkel, L.L. Plant Community Richness Mediates Inhibitory Interactions and Resource Competition between Streptomyces and Fusarium Populations in the Rhizosphere. Microb. Ecol. 2017, 74, 157–167.
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere Microbiome Assemblage Is Affected by Plant Development. ISME J. 2014, 8, 790–803.
- Zhao, M.; Zhao, J.; Yuan, J.; Hale, L.; Wen, T.; Huang, Q.; Vivanco, J.M.; Zhou, J.; Kowalchuk, G.A.; Shen, Q. Root Exudates Drive Soil-microbe-nutrient Feedbacks in Response to Plant Growth. Plant. Cell Environ. 2021, 44, 613–628.
- Stringlis, I.A.; Yu, K.; Feussner, K.; De Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-Dependent Coumarin Exudation Shapes Root Microbiome Assembly to Promote Plant Health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222.
- Harbort, C.J.; Hashimoto, M.; Inoue, H.; Niu, Y.; Guan, R.; Rombolà, A.D.; Kopriva, S.; Voges, M.J.E.E.E.; Sattely, E.S.; Garrido-Oter, R.; et al. Root-Secreted Coumarins and the Microbiota Interact to Improve Iron Nutrition in Arabidopsis. Cell Host Microbe 2020, 28, 825–837.e6.
- Hartmann, M.; Six, J. Soil Structure and Microbiome Functions in Agroecosystems. Nat. Rev. Earth Environ. 2023, 4, 4–18.
- Wang, W.; Jia, T.; Qi, T.; Li, S.; Degen, A.A.; Han, J.; Bai, Y.; Zhang, T.; Qi, S.; Huang, M.; et al. Root Exudates Enhanced Rhizobacteria Complexity and Microbial Carbon Metabolism of Toxic Plants. iScience 2022, 25, 105243.
- Wan, W.; Tan, J.; Wang, Y.; Qin, Y.; He, H.; Wu, H.; Zuo, W.; He, D. Responses of the Rhizosphere Bacterial Community in Acidic Crop Soil to pH: Changes in Diversity, Composition, Interaction, and Function. Sci. Total Environ. 2020, 700, 134418.
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root Exudates: From Plant to Rhizosphere and Beyond. Plant Cell Rep. 2020, 39, 3–17.
- Guo, Y.; Song, B.; Li, A.; Wu, Q.; Huang, H.; Li, N.; Yang, Y.; Adams, J.M.; Yang, L. Higher pH Is Associated with Enhanced Co-Occurrence Network Complexity, Stability and Nutrient Cycling Functions in the Rice Rhizosphere Microbiome. Environ. Microbiol. 2022, 24, 6200–6219.
- Chai, Y.N.; Schachtman, D.P. Root Exudates Impact Plant Performance under Abiotic Stress. Trends Plant Sci. 2022, 27, 80–91.
- Xiong, Y.W.; Li, X.W.; Wang, T.T.; Gong, Y.; Zhang, C.M.; Xing, K.; Qin, S. Root Exudates-Driven Rhizosphere Recruitment of the Plant Growth-Promoting Rhizobacterium Bacillus flexus KLBMP 4941 and Its Growth-Promoting Effect on the Coastal Halophyte Limonium Sinense under Salt Stress. Ecotoxicol. Environ. Saf. 2020, 194, 110374.
- Zadel, U.; Cruzeiro, C.; Raj Durai, A.C.; Nesme, J.; May, R.; Balázs, H.; Michalke, B.; Płaza, G.; Schröder, P.; Schloter, M.; et al. Exudates from Miscanthus x giganteus Change the Response of a Root-Associated Pseudomonas putida Strain towards Heavy Metals. Environ. Pollut. 2022, 313, 119989.
- Feng, F.; Zhan, H.; Wan, Q.; Wang, Y.; Li, Y.; Ge, J.; Sun, X.; Zhu, H.; Yu, X. Rice Recruits Sphingomonas Strain HJY- Rfp via Root Exudate Regulation to Increase Chlorpyrifos Tolerance and Boost Residual Catabolism. J. Exp. Bot. 2021, 72, 5673–5686.
- Williams, A.; de Vries, F.T. Plant Root Exudation under Drought: Implications for Ecosystem Functioning. New Phytol. 2020, 225, 1899–1905.
- Xu, L.; Naylor, D.; Dong, Z.; Simmons, T.; Pierroz, G.; Hixson, K.K.; Kim, Y.M.; Zink, E.M.; Engbrecht, K.M.; Wang, Y.; et al. Drought Delays Development of the Sorghum Root Microbiome and Enriches for Monoderm Bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E4284–E4293.
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere Microbiome Mediates Systemic Root Metabolite Exudation by Root-to-Root Signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883.
- Verbon, E.H.; Liberman, L.M.; Zhou, J.; Yin, J.; Pieterse, C.M.J.; Benfey, P.N.; Stringlis, I.A.; de Jonge, R. Cell-Type-Specific Transcriptomics Reveals That Root Hairs and Endodermal Barriers Play Important Roles in Beneficial Plant-Rhizobacterium Interactions. Mol. Plant 2023, 16, 1160–1177.
- Liu, Z.; Beskrovnaya, P.; Melnyk, R.A.; Hossain, S.S.; Khorasani, S.; O’sullivan, L.R.; Wiesmann, C.L.; Bush, J.; Richard, J.D.; Haney, C.H. A Genome-Wide Screen Identifies Genes in Rhizosphere-Associated Pseudomonas Required to Evade Plant Defenses. mBio 2018, 9, e00433-18.
- Zhang, H.; Liu, Y.; Wu, G.; Dong, X.; Xiong, Q.; Chen, L.; Xu, Z.; Feng, H.; Zhang, R. Bacillus velezensis Tolerance to the Induced Oxidative Stress in Root Colonization Contributed by the Two-component Regulatory System Sensor ResE. Plant. Cell Environ. 2021, 44, 3094–3102.
- Ray, S.; Mishra, S.; Bisen, K.; Singh, S.; Sarma, B.K.; Singh, H.B. Modulation in Phenolic Root Exudate Profile of Abelmoschus esculentus Expressing Activation of Defense Pathway. Microbiol. Res. 2018, 207, 100–107.
- Ankati, S.; Podile, A.R. Metabolites in the Root Exudates of Groundnut Change during Interaction with Plant Growth Promoting Rhizobacteria in a Strain-Specific Manner. J. Plant Physiol. 2019, 243, 153057.
- Schulz-Bohm, K.; Gerards, S.; Hundscheid, M.; Melenhorst, J.; De Boer, W.; Garbeva, P. Calling from Distance: Attraction of Soil Bacteria by Plant Root Volatiles. ISME J. 2018, 12, 1252–1262.
- Dang, K.; Hou, J.; Liu, H.; Peng, J.; Sun, Y.; Li, J.; Dong, Y. Root Exudates of Ginger Induced by Ralstonia solanacearum Infection Could Inhibit Bacterial Wilt. J. Agric. Food Chem. 2023, 71, 1957–1969.
- Hoysted, G.A.; Bell, C.A.; Lilley, C.J.; Urwin, P.E. Aphid Colonization Affects Potato Root Exudate Composition and the Hatching of a Soil Borne Pathogen. Front. Plant Sci. 2018, 9, 1278.
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. Salicylic Acid Modulates Colonization of the Root Microbiome by Specific Bacterial Taxa. Science 2015, 349, 860–864.
More
