Many mechanisms of cancer progression have been explained by principles of ecology, developmental biology, and evolutionary paradigms. Many authors have discussed ecological, developmental, and evolutionary strategies for more successful anti-cancer therapies, or for understanding the ecological, developmental, and evolutionary bases of breast cancer (BC) exploitable vulnerabilities. Researchers use the integrated framework of three well known ecological theories: the Bronfenbrenner’s theory of human development, the Vannote’s River Continuum Concept (RCC), and the Ecological Evolutionary Developmental Biology (Eco-Evo-Devo) theory, to explain and understand several eco-evo-devo-based principles that govern BC progression.
- breast cancer (BC)
- onco-breastomics
- eco-evo-devo theories
- tumorigenesis
- progression
1. BC Is a Hyphenated Eco-Evo-Devo Disease
2. Can Ecological Evolutionary Developmental Biology (Eco-Evo-Devo) Theory Be Applied to BC Development?
3. Bronfenbrenner’s Theory
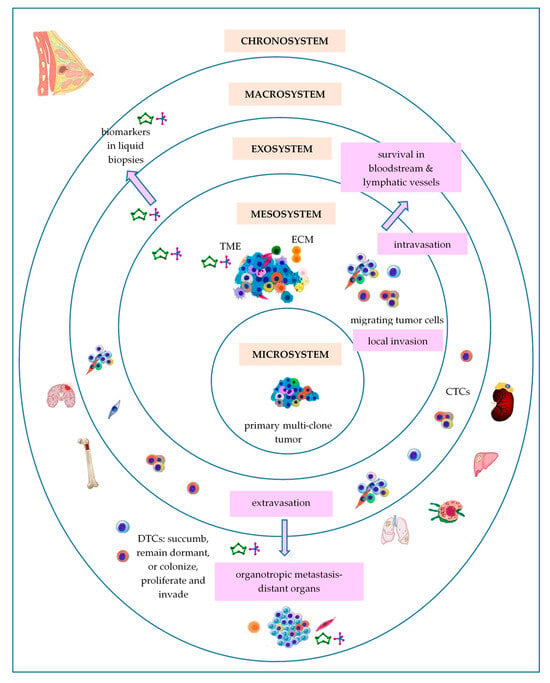
3.1. Primary Breast Tumor as a Microsystem
3.2. Breast Tumor as a Mesosystem
3.3. Breast Tumor as an Exosystem
3.4. Breast Tumor as a Macrosystem
3.5. Breast Tumor as a Chronosystem
4. Vannote’s River Continuum Concept (RCC)
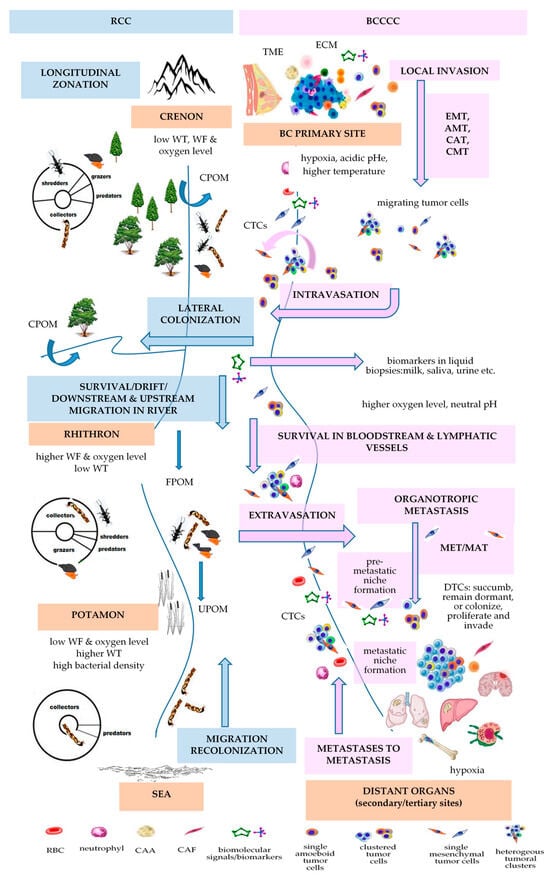
| RCC | [112] | BCCCC and BCPCC | [9] | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Longitudinal Changes in the Benthic Communities in Temperate Rivers | Longitudinal Changes in Kinetics of Metastasis | ||||||||||||||
| ecological zonation | gradient of physical variables [141] | gradient of biological communities | gradient of energy input | BC progression | gradient of physical variables | BCCCC | BCPCC | ||||||||
| headwaters or crenon | water temperature, flow, and oxygen level are low | shredders, collectors, grazers, predators |
CPOM | primary breast tumor | PO2: 2.5–28 mm Hg (mv 10 mm Hg) [70]/<0.1–5% [71]; MR: 100,000–1,200,000 W/m3 [73]; temperature with 1.79 ± 0.88 °C higher that of the surrounding tissue [74]; pHi: 7.3–7.6 & pHe: 6.5–6.9 [77] |
tumor cells; stromal cells (CAFs, TECs, TAPs, CAAs); immune cells (TAMs, TAMCs, TANs, TALs, TAPs, MDSCs); surrounding normal cells (luminal and myoepithelial cells); ECM |
stem-like markers (CD44high/CD24low, EpCAM, PI3K, ALDH1+) | ||||||||
| MIGRATION/DRIFT/COLONIZATION | EMT/INTRAVASATION | ||||||||||||||
| rhithron | high water current and dissolved oxygen; low temperature |
collectors, grazers, shredders, predators | FPOM, UPOM | bloodstream or lymphatic vessels | blood: oxygen level 12% [71] | CTCs | epithelial markers (EpCAM, E-cadherin, CKs, ZO, ESPR1); mesenchymal-like markers (N-cadherin, VIM, Twist1, AKT and PI3K, ZEB1); stemness-like markers (ALDH1, CD44, gangliosides, ABC proteins) [93] |
||||||||
| MIGRATION/DRIFT/COLONIZATION | ETRAVASATION/MET/COLONIZATION | ||||||||||||||
| potamon | low speed; low oxygen content; sandy river bed; higher water temperature; higher bacterial density |
collectors, predators | FPOM, UPOM | preferred BC distant metastatic sites | bone: oxygen levels <1–6% (7–43 mmHg) [105] | homing and dormant DTCs | overexpression of epithelial markers (E-cadherin, occludin, crumbs3); downregulation of mesenchymal markers [142] | ||||||||
References
- Casás-Selves, M.; Degregori, J. How cancer shapes evolution and how evolution shapes cancer. Evol. Educ. Outreach 2011, 4, 624–634.
- Maruki, T.; Ye, Z.; Lynch, M. Evolutionary Genomics of a Subdivided Species. Mol. Biol. Evol. 2022, 39, msac152.
- Frank, S.A. Somatic evolutionary genomics: Mutations during development cause highly variable genetic mosaicism with risk of cancer and neurodegeneration. Proc. Natl. Acad. Sci. USA 2010, 107 (Suppl. S1), 1725–1730.
- Baer, B.; Millar, A.H. Proteomics in evolutionary ecology. J. Proteom. 2016, 135, 4–11.
- Reynolds, B.A.; Oli, M.W.; Oli, M.K. Eco-oncology: Applying ecological principles to understand and manage cancer. Ecol. Evol. 2020, 10, 8538–8553.
- Kareva, I. Cancer Ecology: Niche Construction, Keystone Species, Ecological Succession, and Ergodic Theory. Biol. Theory 2015, 10, 283–288.
- Chen, X.; Song, E. The theory of tumor ecosystem. Cancer Commun. 2022, 42, 587–608.
- Somarelli, J. The Hallmarks of Cancer as Ecologically Driven Phenotypes. Front. Ecol. Evol. 2021, 9, 661583.
- Neagu, A.-N.; Whitham, D.; Buonanno, E.; Jenkins, A.; Alexa-Stratulat, T.; Tamba, B.I.; Darie, C.C. Proteomics and its applications in breast cancer. Am. J. Cancer Res. 2021, 11, 4006–4049.
- Boddy, A.M. The need for evolutionary theory in cancer research. Eur. J. Epidemiol. 2022, 38, 1259–1264.
- Coscieme, L.; Pulselli, F.M.; Jørgensen, S.E.; Bastianoni, S.; Marchettini, N. Thermodynamics-based categorization of ecosystems in a socio-ecological context. Ecol. Model. 2013, 258, 1–8.
- Modaresi Movahed, T.; Jalaly Bidgoly, H.; Khoshgoftar Manesh, M.H.; Mirzaei, H.R. Predicting cancer cells progression via entropy generation based on AR and ARMA models. Int. Commun. Heat Mass Transf. 2021, 127, 105565.
- Myers, K.V.; Pienta, K.J.; Amend, S.R. Cancer Cells and M2 Macrophages: Cooperative Invasive Ecosystem Engineers. Cancer Control J. Moffitt Cancer Cent. 2020, 27, 1073274820911058.
- Sell, S.; Nicolini, A.; Ferrari, P.; Biava, P.M. Cancer: A Problem of Developmental Biology; Scientific Evidence for Reprogramming and Differentiation Therapy. Curr. Drug Targets 2016, 17, 1103–1110.
- Edwards, P.A.W. The Impact of Developmental Biology on Cancer Research: An Overview. Cancer Metastasis Rev. 1999, 18, 175–180.
- Zhu, X.; Li, S.; Xu, B.; Luo, H. Cancer evolution: A means by which tumors evade treatment. Biomed. Pharmacother. 2021, 133, 111016.
- Thomas, F.; Fisher, D.; Fort, P.; Marie, J.-P.; Daoust, S.; Roche, B.; Grunau, C.; Cosseau, C.; Mitta, G.; Baghdiguian, S.; et al. Applying ecological and evolutionary theory to cancer: A long and winding road. Evol. Appl. 2013, 6, 1–10.
- Chouaib, S.; Lorens, J. Editorial: Targeting the Tumor Microenvironment for a More Effective and Efficient Cancer Immunotherapy. Front. Immunol. 2020, 11, 933.
- Kotler, B.P.; Brown, J.S. Cancer Community Ecology. Cancer Control J. Moffitt Cancer Cent. 2020, 27, 1073274820951776.
- Dujon, A.M.; Aktipis, A.; Alix-Panabières, C.; Amend, S.R.; Boddy, A.M.; Brown, J.S.; Capp, J.-P.; DeGregori, J.; Ewald, P.; Gatenby, R.; et al. Identifying key questions in the ecology and evolution of cancer. Evol. Appl. 2021, 14, 877–892.
- Tot, T. Breast Cancer: A Lobar Disease; Springer: London, UK, 2008; pp. 1–216.
- Tot, T. The Theory of the Sick Breast Lobe and the Possible Consequences. Int. J. Surg. Pathol. 2007, 15, 369–375.
- Tot, T. DCIS, cytokeratins, and the theory of the sick lobe. Virchows Arch. 2005, 447, 1–8.
- Tan, M.; Tot, T. The sick lobe hypothesis, field cancerisation and the new era of precision breast surgery. Gland. Surg. 2018, 7, 611–618.
- Li, L.; Sullivan, P.L.; Benettin, P.; Cirpka, O.A.; Bishop, K.; Brantley, S.L.; Knapp, J.L.A.; van Meerveld, I.; Rinaldo, A.; Seibert, J.; et al. Toward catchment hydro-biogeochemical theories. WIREs Water 2021, 8, e1495.
- Jiménez-Navarro, I.C.; Mesman, J.P.; Pierson, D.; Trolle, D.; Nielsen, A.; Senent-Aparicio, J. Application of an integrated catchment-lake model approach for simulating effects of climate change on lake inputs and biogeochemistry. Sci. Total Environ. 2023, 885, 163946.
- Chroni, A.; Miura, S.; Oladeinde, O.; Aly, V.; Kumar, S. Migrations of cancer cells through the lens of phylogenetic biogeography. Sci. Rep. 2021, 11, 17184.
- Chroni, A.; Kumar, S. Tumors Are Evolutionary Island-Like Ecosystems. Genome Biol. Evol. 2021, 13, evab276.
- Gatenby, R.A.; Artzy-Randrup, Y.; Epstein, T.; Reed, D.R.; Brown, J.S. Eradicating Metastatic Cancer and the Eco-Evolutionary Dynamics of Anthropocene Extinctions. Cancer Res. 2020, 80, 613–623.
- Miller, A.K.; Brown, J.S.; Enderling, H.; Basanta, D.; Whelan, C.J. The Evolutionary Ecology of Dormancy in Nature and in Cancer. Front. Ecol. Evol. 2021, 9, 676802.
- Gatenbee, C.D.; Minor, E.S.; Slebos, R.J.C.; Chung, C.H.; Anderson, A.R.A. Histoecology: Applying Ecological Principles and Approaches to Describe and Predict Tumor Ecosystem Dynamics Across Space and Time. Cancer Control J. Moffitt Cancer Cent. 2020, 27, 1073274820946804.
- Noorbakhsh, J.; Zhao, Z.-M.; Russell, J.C.; Chuang, J.H. Treating Cancer as an Invasive Species. Mol. Cancer Res. MCR 2020, 18, 20–26.
- Neinavaie, F.; Ibrahim-Hashim, A.; Kramer, A.M.; Brown, J.S.; Richards, C.L. The Genomic Processes of Biological Invasions: From Invasive Species to Cancer Metastases and Back Again. Front. Ecol. Evol. 2021, 9, 681100.
- Pouliquen, D.L.; Boissard, A.; Coqueret, O.; Guette, C. Biomarkers of tumor invasiveness in proteomics (Review). Int. J. Oncol. 2020, 57, 409–432.
- Bi, G.; Liang, J.; Zheng, Y.; Li, R.; Zhao, M.; Huang, Y.; Zhan, C.; Xu, S.; Fan, H. Multi-omics characterization and validation of invasiveness-related molecular features across multiple cancer types. J. Transl. Med. 2021, 19, 124.
- Cortés-Hernández, L.E.; Eslami-S, Z.; Dujon, A.M.; Giraudeau, M.; Ujvari, B.; Thomas, F.; Alix-Panabières, C. Do malignant cells sleep at night? Genome Biol. 2020, 21, 276.
- Arnal, A.; Beckmann, C.; Biro, P.A.; Boidin-Wichlacz, C.; Massol, F.; Mery, F.; Misse, D.; Poulin, R.; Renaud, F.; Roche, B.; et al. Cancer and life-history traits: Lessons from host–parasite interactions. Parasitology 2016, 143, 533–541.
- Gerstung, M.; Jolly, C.; Leshchiner, I.; Dentro, S.C.; Gonzalez, S.; Rosebrock, D.; Mitchell, T.J.; Rubanova, Y.; Anur, P.; Yu, K.; et al. The evolutionary history of 2658 cancers. bioRxiv 2017, 161562.
- Fortunato, A.; Boddy, A.; Mallo, D.; Aktipis, A.; Maley, C.C.; Pepper, J.W. Natural Selection in Cancer Biology: From Molecular Snowflakes to Trait Hallmarks. Cold Spring Harb. Perspect. Med. 2017, 7, a029652.
- Wang, J.; Khiabanian, H.; Rossi, D.; Fabbri, G.; Gattei, V.; Forconi, F.; Laurenti, L.; Marasca, R.; Del Poeta, G.; Foà, R.; et al. Tumor evolutionary directed graphs and the history of chronic lymphocytic leukemia. eLife 2014, 3, e02869.
- Vineis, P. Cancer as an evolutionary process at the cell level: An epidemiological perspective. Carcinogenesis 2003, 24, 1–6.
- Alfarouk, K.O.; Ibrahim, M.E.; Gatenby, R.A.; Brown, J.S. Riparian ecosystems in human cancers. Evol. Appl. 2013, 6, 46–53.
- Seferbekova, Z.; Lomakin, A.; Yates, L.R.; Gerstung, M. Spatial biology of cancer evolution. Nat. Rev. Genet. 2023, 24, 295–313.
- Gilbert, S.F.; Bosch, T.C.G.; Ledón-Rettig, C. Eco-Evo-Devo: Developmental symbiosis and developmental plasticity as evolutionary agents. Nat. Rev. Genet. 2015, 16, 611–622.
- Abouheif, E.; Favé, M.-J.; Ibarrarán-Viniegra, A.S.; Lesoway, M.P.; Rafiqi, A.M.; Rajakumar, R. Eco-Evo-Devo: The Time Has Come. In Ecological Genomics: Ecology and the Evolution of Genes and Genomes; Landry, C.R., Aubin-Horth, N., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 107–125.
- Sultan, S.E. Eco-Evo-Devo. In Evolutionary Developmental Biology: A Reference Guide; Nuno de la Rosa, L., Müller, G., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–13.
- Plowman, P.N.; Plowman, C.E. Onco-ontogeny recapitulates phylogeny–A consideration. Oncogene 2021, 40, 1542–1550.
- Liu, W.; Deng, Y.; Li, Z.; Chen, Y.; Zhu, X.; Tan, X.; Cao, G. Cancer Evo–Dev: A Theory of Inflammation-Induced Oncogenesis. Front. Immunol. 2021, 12, 768098.
- Kozlov, A.P. Carcino-Evo-Devo, A Theory of the Evolutionary Role of Hereditary Tumors. Int. J. Mol. Sci. 2023, 24, 8611.
- Thomas, F.; Jacqueline, C.; Tissot, T.; Henard, M.; Blanchet, S.; Loot, G.; Dawson, E.; Mery, F.; Renaud, F.; Montagne, J.; et al. The importance of cancer cells for animal evolutionary ecology. Nat. Ecol. Evol. 2017, 1, 1592–1595.
- Rosa, E.M.; Tudge, J. Urie Bronfenbrenner’s Theory of Human Development: Its Evolution from Ecology to Bioecology. J. Fam. Theory Rev. 2013, 5, 243–258.
- Pask, S.; Pinto, C.; Bristowe, K.; van Vliet, L.; Nicholson, C.; Evans, C.J.; George, R.; Bailey, K.; Davies, J.M.; Guo, P.; et al. A framework for complexity in palliative care: A qualitative study with patients, family carers and professionals. Palliat. Med. 2018, 32, 1078–1090.
- Yeremeyev, I.; Dychko, A.; Remez, N.; Kraychuk, S.; Ostapchuk, N. Problems of sustainable development of ecosystems. IOP Conf. Ser. Earth Environ. Sci. 2021, 628, 012014.
- Majumdar, S.; Liu, S.-T. Cell division symmetry control and cancer stem cells. AIMS Mol. Sci. 2020, 7, 82–98.
- Zhang, X.; Powell, K.; Li, L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers 2020, 12, 3765.
- Nalla, L.V.; Kalia, K.; Khairnar, A. Self-renewal signaling pathways in breast cancer stem cells. Int. J. Biochem. Cell Biol. 2019, 107, 140–153.
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238.
- Heyde, A.; Reiter, J.G.; Naxerova, K.; Nowak, M.A. Consecutive seeding and transfer of genetic diversity in metastasis. Proc. Natl. Acad. Sci. USA 2019, 116, 14129–14137.
- Nishimura, T.; Kakiuchi, N.; Yoshida, K.; Sakurai, T.; Kataoka, T.R.; Kondoh, E.; Chigusa, Y.; Kawai, M.; Sawada, M.; Inoue, T.; et al. Evolutionary histories of breast cancer and related clones. Nature 2023, 620, 607–614.
- Miller, W. The Hierarchical Structure of Ecosystems: Connections to Evolution. Evol. Educ. Outreach 2008, 1, 16–24.
- Cole, A.J.; Fayomi, A.P.; Anyaeche, V.I.; Bai, S.; Buckanovich, R.J. An evolving paradigm of cancer stem cell hierarchies: Therapeutic implications. Theranostics 2020, 10, 3083–3098.
- Pelage, L.; Lucena-Frédou, F.; Eduardo, L.N.; Le Loc’h, F.; Bertrand, A.; Lira, A.S.; Frédou, T. Competing with each other: Fish isotopic niche in two resource availability contexts. Front. Mar. Sci. 2022, 9, 975091.
- Johnston, L.A. Socializing with MYC: Cell competition in development and as a model for premalignant cancer. Cold Spring Harb. Perspect. Med. 2014, 4, a014274.
- Nicolazzo, C.; Francescangeli, F.; Magri, V.; Giuliani, A.; Zeuner, A.; Gazzaniga, P. Is cancer an intelligent species? Cancer Metastasis Rev. 2023, 42, 1201–1218.
- Madan, E.; Palma, A.M.; Vudatha, V.; Trevino, J.G.; Natarajan, K.N.; Winn, R.A.; Won, K.J.; Graham, T.A.; Drapkin, R.; McDonald, S.A.C.; et al. Cell Competition in Carcinogenesis. Cancer Res. 2022, 82, 4487–4496.
- Desjardins-Lecavalier, N.; Annis, M.G.; Nowakowski, A.; Kiepas, A.; Binan, L.; Roy, J.; Modica, G.; Hébert, S.; Kleinman, C.L.; Siegel, P.M.; et al. Migration speed of captured breast cancer subpopulations correlates with metastatic fitness. J. Cell Sci. 2023, 136, jcs260835.
- Angeler, D.G.; Fried-Petersen, H.B.; Allen, C.R.; Garmestani, A.; Twidwell, D.; Chuang, W.-C.; Donovan, V.M.; Eason, T.; Roberts, C.P.; Sundstrom, S.M.; et al. Chapter One–Adaptive capacity in ecosystems. In Advances in Ecological Research; Bohan, D.A., Dumbrell, A.J., Eds.; Academic Press: New York, NY, USA, 2019; Volume 60, pp. 1–24.
- Dong, X.; Fisher, S.G. Ecosystem spatial self-organization: Free order for nothing? Ecol. Complex. 2019, 38, 24–30.
- Balcioglu, O.; Heinz, R.E.; Freeman, D.W.; Gates, B.L.; Hagos, B.M.; Booker, E.; Mirzaei Mehrabad, E.; Diesen, H.T.; Bhakta, K.; Ranganathan, S.; et al. CRIPTO antagonist ALK4L75A-Fc inhibits breast cancer cell plasticity and adaptation to stress. Breast Cancer Res. 2020, 22, 125.
- Gilkes, D.M.; Semenza, G.L. Role of hypoxia-inducible factors in breast cancer metastasis. Future Oncol. 2013, 9, 1623–1636.
- Campillo, N.; Falcones, B.; Otero, J.; Colina, R.; Gozal, D.; Navajas, D.; Farré, R.; Almendros, I. Differential Oxygenation in Tumor Microenvironment Modulates Macrophage and Cancer Cell Crosstalk: Novel Experimental Setting and Proof of Concept. Front. Oncol. 2019, 9, 43.
- García-Jiménez, C.; Goding, C.R. Starvation and Pseudo-Starvation as Drivers of Cancer Metastasis through Translation Reprogramming. Cell Metab. 2019, 29, 254–267.
- Lozano, A., 3rd; Hayes, J.C.; Compton, L.M.; Azarnoosh, J.; Hassanipour, F. Determining the thermal characteristics of breast cancer based on high-resolution infrared imaging, 3D breast scans, and magnetic resonance imaging. Sci. Rep. 2020, 10, 10105.
- Yahara, T.; Koga, T.; Yoshida, S.; Nakagawa, S.; Deguchi, H.; Shirouzu, K. Relationship between Microvessel Density and Thermographic Hot Areas in Breast Cancer. Surg. Today 2003, 33, 243–248.
- Rolver, M.G.; Pedersen, S.F. Putting Warburg to work: How imaging of tumour acidosis could help predict metastatic potential in breast cancer. Br. J. Cancer 2021, 124, 1–2.
- White, K.A.; Grillo-Hill, B.K.; Barber, D.L. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J. Cell Sci. 2017, 130, 663–669.
- Ibrahim-Hashim, A.; Estrella, V. Acidosis and cancer: From mechanism to neutralization. Cancer Metastasis Rev. 2019, 38, 149–155.
- Greaves, M.; Maley, C.C. Clonal evolution in cancer. Nature 2012, 481, 306–313.
- Li, J.J.; Tsang, J.Y.; Tse, G.M. Tumor Microenvironment in Breast Cancer–Updates on Therapeutic Implications and Pathologic Assessment. Cancers 2021, 13, 4233.
- Mayer, S.; Milo, T.; Isaacson, A.; Halperin, C.; Miyara, S.; Stein, Y.; Lior, C.; Pevsner-Fischer, M.; Tzahor, E.; Mayo, A.; et al. The tumor microenvironment shows a hierarchy of cell-cell interactions dominated by fibroblasts. Nat. Commun. 2023, 14, 5810.
- Hu, D.; Li, Z.; Zheng, B.; Lin, X.; Pan, Y.; Gong, P.; Zhuo, W.; Hu, Y.; Chen, C.; Chen, L.; et al. Cancer-associated fibroblasts in breast cancer: Challenges and opportunities. Cancer Commun. 2022, 42, 401–434.
- Takebe, N.; Ivy, P.; Timmer, W.; Khan, M.; Schulz; Harris, P. Cancer-associated Fibroblasts and Therapies That Interfere with Their Activity. Tumor Microenviron. Ther. 2013, 1, 19–36.
- Kareva, I.; Luddy, K.A.; O’Farrelly, C.; Gatenby, R.A.; Brown, J.S. Predator-Prey in Tumor-Immune Interactions: A Wrong Model or Just an Incomplete One? Front. Immunol. 2021, 12, 668221.
- Taylor, T.B.; Wass, A.V.; Johnson, L.J.; Dash, P. Resource competition promotes tumour expansion in experimentally evolved cancer. BMC Evol. Biol. 2017, 17, 268.
- Chang, C.-H.; Qiu, J.; O’Sullivan, D.; Buck, M.D.; Noguchi, T.; Curtis, J.D.; Chen, Q.; Gindin, M.; Gubin, M.M.; van der Windt, G.J.W.; et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 2015, 162, 1229–1241.
- Brown, R.A.M.; Richardson, K.L.; Kabir, T.D.; Trinder, D.; Ganss, R.; Leedman, P.J. Altered Iron Metabolism and Impact in Cancer Biology, Metastasis, and Immunology. Front. Oncol. 2020, 10, 476.
- Bañuls, A.-L.; Thomas, F.; Renaud, F. Of parasites and men. Infect. Genet. Evol. 2013, 20, 61–70.
- Song, X.; Wei, C.; Li, X. The Relationship between Microbial Community and Breast Cancer. Front. Cell Infect. Microbiol. 2022, 12, 849022.
- Liao, W.-T.; Ye, Y.-P.; Deng, Y.-J.; Bian, X.-W.; Ding, Y.-Q. Metastatic cancer stem cells: From the concept to therapeutics. Am. J. Stem Cells 2014, 3, 46–62.
- Baumann, Z.; Auf der Maur, P.; Bentires-Alj, M. Feed-forward loops between metastatic cancer cells and their microenvironment–The stage of escalation. EMBO Mol. Med. 2022, 14, e14283.
- Somveille, M.; Ellis-Soto, D. Linking animal migration and ecosystem processes: Data-driven simulation of propagule dispersal by migratory herbivores. Ecol. Evol. 2022, 12, e9383.
- Wu, J.-S.; Jiang, J.; Chen, B.-J.; Wang, K.; Tang, Y.-L.; Liang, X.-H. Plasticity of cancer cell invasion: Patterns and mechanisms. Transl. Oncol. 2021, 14, 100899.
- Barriere, G.; Fici, P.; Gallerani, G.; Fabbri, F.; Zoli, W.; Rigaud, M. Circulating tumor cells and epithelial, mesenchymal and stemness markers: Characterization of cell subpopulations. Ann. Transl. Med. 2014, 2, 109.
- Liu, F.; Gu, L.-N.; Shan, B.-E.; Geng, C.-Z.; Sang, M.-X. Biomarkers for EMT and MET in breast cancer: An update. Oncol. Lett. 2016, 12, 4869–4876.
- Sciacovelli, M.; Frezza, C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J. 2017, 284, 3132–3144.
- Miller, K.M.; Schulze, A.D.; Ginther, N.; Li, S.; Patterson, D.A.; Farrell, A.P.; Hinch, S.G. Salmon spawning migration: Metabolic shifts and environmental triggers. Comp. Biochem. Physiol. Part D Genom. Proteom. 2009, 4, 75–89.
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast cancer subtypes predict the preferential site of distant metastases: A SEER based study. Oncotarget 2017, 8, 27990–27996.
- Wu, M.; Liang, Y.; Zhang, X. Changes in Pulmonary Microenvironment Aids Lung Metastasis of Breast Cancer. Front. Oncol. 2022, 12, 860932.
- Lim, S.; Nam, H.; Jeon, J.S. Chemotaxis Model for Breast Cancer Cells Based on Signal/Noise Ratio. Biophys. J. 2018, 115, 2034–2043.
- Farahani, M.K.; Gharibshahian, M.; Rezvani, A.; Vaez, A. Breast cancer brain metastasis: From etiology to state-of-the-art modeling. J. Biol. Eng. 2023, 17, 41.
- Chen, W.; Hoffmann, A.D.; Liu, H.; Liu, X. Organotropism: New insights into molecular mechanisms of breast cancer metastasis. npj Precis. Oncol. 2018, 2, 4.
- Schrijver, W.A.M.E.; Selenica, P.; Lee, J.Y.; Ng, C.K.Y.; Burke, K.A.; Piscuoglio, S.; Berman, S.H.; Reis-Filho, J.S.; Weigelt, B.; van Diest, P.J.; et al. Mutation Profiling of Key Cancer Genes in Primary Breast Cancers and Their Distant Metastases. Cancer Res. 2018, 78, 3112–3121.
- Yumoto, K.; Eber, M.R.; Berry, J.E.; Taichman, R.S.; Shiozawa, Y. Molecular pathways: Niches in metastatic dormancy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 3384–3389.
- Măgălie, A.; Schwartz, D.A.; Lennon, J.T.; Weitz, J.S. Optimal dormancy strategies in fluctuating environments given delays in phenotypic switching. J. Theor. Biol. 2023, 561, 111413.
- Johnson, R.W.; Sowder, M.E.; Giaccia, A.J. Hypoxia and Bone Metastatic Disease. Curr. Osteoporos. Rep. 2017, 15, 231–238.
- Bushnell, G.G.; Deshmukh, A.P.; den Hollander, P.; Luo, M.; Soundararajan, R.; Jia, D.; Levine, H.; Mani, S.A.; Wicha, M.S. Breast cancer dormancy: Need for clinically relevant models to address current gaps in knowledge. npj Breast Cancer 2021, 7, 66.
- Tivari, S.; Lu, H.; Dasgupta, T.; De Lorenzo, M.S.; Wieder, R. Reawakening of dormant estrogen-dependent human breast cancer cells by bone marrow stroma secretory senescence. Cell Commun. Signal. 2018, 16, 48.
- Bleicher, R.J. Timing and Delays in Breast Cancer Evaluation and Treatment. Ann. Surg. Oncol. 2018, 25, 2829–2838.
- Wu, J.; Liu, H.; Hu, T.; Long, M.; Zhou, X.; Wang, S. The natural history of breast cancer: A chronological analysis of breast cancer progression using data from the SEER database. Ann. Transl. Med. 2022, 10, 365.
- Hu, Z.; Curtis, C. Looking backward in time to define the chronology of metastasis. Nat. Commun. 2020, 11, 3213.
- Menes, T.S.; Terry, M.B.; Goldgar, D.; Andrulis, I.L.; Knight, J.A.; John, E.M.; Liao, Y.; Southey, M.; Miron, A.; Chung, W.; et al. Second primary breast cancer in BRCA1 and BRCA2 mutation carriers: 10-year cumulative incidence in the Breast Cancer Family Registry. Breast Cancer Res. Treat. 2015, 151, 653–660.
- Vannote, R.L.; Minshall, G.W.; Cummins, K.W.; Sedell, J.R.; Gushing, C.E. The River Continuum Concept. Can. J. Fish. Aquat. Sci. 1980, 37, 130–137.
- Doretto, A.; Piano, E.; Larson, C.E. The River Continuum Concept: Lessons from the past and perspectives for the future. Can. J. Fish. Aquat. Sci. 2020, 77, 1853–1864.
- Tinganelli, W.; Durante, M. Tumor Hypoxia and Circulating Tumor Cells. Int. J. Mol. Sci. 2020, 21, 9592.
- Rao, S.M.N.; Tata, U.; Lin, V.K.; Chiao, J.-C. The Migration of Cancer Cells in Gradually Varying Chemical Gradients and Mechanical Constraints. Micromachines 2014, 5, 13–26.
- Garg, A.A.; Jones, T.H.; Moss, S.M.; Mishra, S.; Kaul, K.; Ahirwar, D.K.; Ferree, J.; Kumar, P.; Subramaniam, D.; Ganju, R.K.; et al. Electromagnetic fields alter the motility of metastatic breast cancer cells. Commun. Biol. 2019, 2, 303.
- Varennes, J.; Moon, H.-R.; Saha, S.; Mugler, A.; Han, B. Physical constraints on accuracy and persistence during breast cancer cell chemotaxis. PLoS Comput. Biol. 2019, 15, e1006961.
- Liu, Z.; Lee, S.J.; Park, S.; Konstantopoulos, K.; Glunde, K.; Chen, Y.; Barman, I. Cancer cells display increased migration and deformability in pace with metastatic progression. FASEB J. 2020, 34, 9307–9315.
- Couzin, I.D. Collective animal migration. Curr. Biol. 2018, 28, R976–R980.
- Amintas, S.; Bedel, A.; Moreau-Gaudry, F.; Boutin, J.; Buscail, L.; Merlio, J.-P.; Vendrely, V.; Dabernat, S.; Buscail, E. Circulating Tumor Cell Clusters: United We Stand Divided We Fall. Int. J. Mol. Sci. 2020, 21, 2653.
- Kubelka, V.; Sandercock, B.K.; Székely, T.; Freckleton, R.P. Animal migration to northern latitudes: Environmental changes and increasing threats. Trends Ecol. Evol. 2022, 37, 30–41.
- Rejniak, K.A. Circulating Tumor Cells: When a Solid Tumor Meets a Fluid Microenvironment. In Systems Biology of Tumor Microenvironment; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2016; Volume 936, pp. 93–106.
- Bulfoni, M.; Turetta, M.; Del Ben, F.; Di Loreto, C.; Beltrami, A.P.; Cesselli, D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. Int. J. Mol. Sci. 2016, 17, 1775.
- Menyailo, M.E.; Tretyakova, M.S.; Denisov, E.V. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. Int. J. Mol. Sci. 2020, 21, 1696.
- Heitzer, E.; Auer, M.; Gasch, C.; Pichler, M.; Ulz, P.; Hoffmann, E.M.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Lackner, C.; et al. Complex Tumor Genomes Inferred from Single Circulating Tumor Cells by Array-CGH and Next-Generation Sequencing. Cancer Res. 2013, 73, 2965–2975.
- Park, H.-A.; Brown, S.R.; Kim, Y. Cellular Mechanisms of Circulating Tumor Cells During Breast Cancer Metastasis. Int. J. Mol. Sci. 2020, 21, 5040.
- Shin, D.; Park, J.; Han, D.; Moon, J.H.; Ryu, H.S.; Kim, Y. Identification of TUBB2A by quantitative proteomic analysis as a novel biomarker for the prediction of distant metastatic breast cancer. Clin. Proteom. 2020, 17, 16.
- Veyssière, H.; Bidet, Y.; Penault-Llorca, F.; Radosevic-Robin, N.; Durando, X. Circulating proteins as predictive and prognostic biomarkers in breast cancer. Clin. Proteom. 2022, 19, 25.
- Kjølle, S.; Finne, K.; Birkeland, E.; Ardawatia, V.; Winge, I.; Aziz, S.; Knutsvik, G.; Wik, E.; Paulo, J.A.; Vethe, H.; et al. Hypoxia induced responses are reflected in the stromal proteome of breast cancer. Nat. Commun. 2023, 14, 3724.
- Yates, L.R.; Gerstung, M.; Knappskog, S.; Desmedt, C.; Gundem, G.; Van Loo, P.; Aas, T.; Alexandrov, L.B.; Larsimont, D.; Davies, H.; et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 2015, 21, 751–759.
- Fernandez-Garcia, D.; Nteliopoulos, G.; Hastings, R.K.; Rushton, A.; Page, K.; Allsopp, R.C.; Ambasager, B.; Gleason, K.; Guttery, D.S.; Ali, S.; et al. Shallow WGS of individual CTCs identifies actionable targets for informing treatment decisions in metastatic breast cancer. Br. J. Cancer 2022, 127, 1858–1864.
- Bhatia, R.; Chang, J.; Munoz, J.L.; Walker, N.D. Forging New Therapeutic Targets: Efforts of Tumor Derived Exosomes to Prepare the Pre-Metastatic Niche for Cancer Cell Dissemination and Dormancy. Biomedicines 2023, 11, 1614.
- Yuan, X.; Qian, N.; Ling, S.; Li, Y.; Sun, W.; Li, J.; Du, R.; Zhong, G.; Liu, C.; Yu, G.; et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics 2021, 11, 1429–1445.
- Chen, K.M.; Stephen, J.K.; Raju, U.; Worsham, M.J. Delineating an Epigenetic Continuum for Initiation, Transformation and Progression to Breast Cancer. Cancers 2011, 3, 1580–1592.
- Sanati, S. Morphologic and Molecular Features of Breast Ductal Carcinoma in Situ. Am. J. Pathol. 2019, 189, 946–955.
- Lüönd, F.; Sugiyama, N.; Bill, R.; Bornes, L.; Hager, C.; Tang, F.; Santacroce, N.; Beisel, C.; Ivanek, R.; Bürglin, T.; et al. Distinct contributions of partial and full EMT to breast cancer malignancy. Dev. Cell 2021, 56, 3203–3221.e11.
- Manfioletti, G.; Fedele, M. Epithelial-Mesenchymal Transition (EMT). Int. J. Mol. Sci. 2023, 24, 11386.
- Tomaskovic-Crook, E.; Thompson, E.W.; Thiery, J.P. Epithelial to mesenchymal transition and breast cancer. Breast Cancer Res. 2009, 11, 213.
- Malagoli Tagliazucchi, G.; Wiecek, A.J.; Withnell, E.; Secrier, M. Genomic and microenvironmental heterogeneity shaping epithelial-to-mesenchymal trajectories in cancer. Nat. Commun. 2023, 14, 789.
- McFaline-Figueroa, J.L.; Hill, A.J.; Qiu, X.; Jackson, D.; Shendure, J.; Trapnell, C. A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition. Nat. Genet. 2019, 51, 1389–1398.
- Kamboj, V.; Kamboj, N.; Sharma, A. A review on general characteristics, classification and degradation of river systems. In Environmental Degradation: Causes and Remediation Strategies; Agro Environ Media, Agriculture and Environmental Science Academy: Haridwar, India, 2020; pp. 47–62.
- Terceiro, L.E.L.; Edechi, C.A.; Ikeogu, N.M.; Nickel, B.E.; Hombach-Klonisch, S.; Sharif, T.; Leygue, E.; Myal, Y. The Breast Tumor Microenvironment: A Key Player in Metastatic Spread. Cancers 2021, 13, 4798.
