NSD3 (nuclear receptor-binding SET domain protein 3 (NSD3) is a member of the NSD histone methyltransferase family of proteins. In recent years, it has been identified as a potential oncogene in certain types of cancer. The NSD3 gene encodes three isoforms, the long version (NSD3L), a short version (NSD3S) and the WHISTLE isoforms. Importantly, the NSD3S isoform corresponds to the N-terminal region of the full-length protein, lacking the methyltransferase domain. The chromosomal location of NSD3 is frequently amplified across cancer types, such as breast, lung, and colon, among others. This amplification has been correlated to a chromothripsis event, that could explain the different NSD3 alterations found in cancer. The fusion proteins containing NSD3 have also been reported in leukemia (NSD3-NUP98), and in NUT (nuclear protein of the testis) midline carcinoma (NSD3-NUT). Its role as an oncogene has been described by modulating different cancer pathways through its methyltransferase activity, or the short isoform of the protein, through protein interactions.
- cancer
- molecular oncology
- oncogenes
- NSD3
- NSD3S
1. Introduction
2. NSD3 Protein Structure
NSD3 was first described in 2000 by studying the PWWP (proline-tryptophan-tryptophan-proline) domain of NSD2 and performing a database search for proteins having the PWWP domain in their structure. The NSD3 gene is found on chromosome 8p11.2 [12] and encodes three isoforms by alternative splicing. The long isoform, termed NSD3L, is a protein of 1437 amino acids [13]. Alternative splicing of exon 10 encodes a protein of 645 amino acids, named NSD3 short (NSD3S), which is identical to NSD3L in the first 619 amino acids [12]. Finally, isoform WHISTLE (WHSC1-like 1 isoform 9 with methyltransferase activity to lysine) is a short alternative splice version of the C- terminal of NSD3L that encodes a protein of 506 amino acids. NSD3L has five PHD (plant-homeodomain)-type zinc fingers motifs, two PWWP domains, and the methyltransferase SET domain. Right next to the SET domain there is a SAC (SET-associated Cys-rich) domain rich in cysteines, followed by a Cys-His-rich domain termed C5HCH motif near the C terminal end of the protein [13,14][13][14]. The PWWP domain is a histone methyl-lysine (H3K36) reader, acting as an epigenetic regulator of gene expression [15[15][16][17],16,17], and has been postulated as a site for protein–protein interactions due to the amino acid composition [18]. The PHD domain binds chromatin at histone H3 lysine 4 unmodified or methylated [19]. The SET domain is a region conserved between the SET family of methyltransferases, with specificity for mono- or di-methylation of H3 lysine 36. The SET domain is separated into three smaller segments, the pre-SET, SET and post-SET domain, all of which are needed for catalytic activity [20].3. NSD3 Alterations in Cancer
NSD3 proteins are ubiquitously expressed in human tissues, with the NSD3S isoform being more prominent than NSD3L [22[21][22][23],23,24], and the WHISTLE isoform primarily found in the testis [25][24]. Compared to NSD1 and NSD2, NSD3 exhibits a higher genetic variation and amplification in cancer. The oncogenic role of NSD3 is manifested by changes ranging from alterations in expression, such as overexpression and point mutations, as well as fusions with other proteins which result in differences in cellular activity (Figure 21).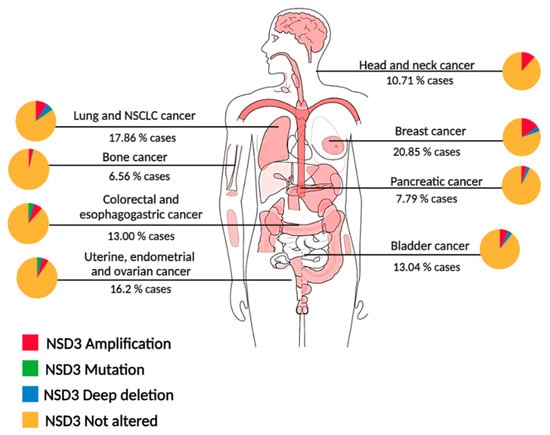
3.1. Study of the Amplicon 8p11-12: Chromothripsis
3.1.1. Amplification
Because of the 8p11-12 amplicon found in different epithelial cancers [33][29], NSD3 has been proposed, among other proteins, as an important oncogene for cancer progression. Using different approaches, such as overexpression of NSD3, small interfering RNA (siRNA) and short hairpin RNA (shRNA)-mediated knockdown against NSD3 in 8p11-12 amplified breast cancer cells, it was found that the loss of NSD3 resulted in a profound loss of the growth and survival of these cells, indicating a function for this protein in regulating survival and transformation [22,34][21][30]. In a breast cancer mouse model expressing NSD3 in the mammary epithelium, NSD3 was revealed as a transforming oncogene by exhibiting mammary hyperplasia, dysplasia, and invasiveness [35][31]. NSD3 has also been proposed as an oncogenic driver in non-small cell lung cancer (NSCLC) [11], lung squamous cell carcinoma (LUSC) [36][32] and pancreatic ductal adenocarcinoma (PDAC) where the 8p11-12 amplicon has also been found. The studies validated the consistent amplification of NSD3 and showed that the depletion of NSD3 decreases the viability and the colony formation capacity of lung and pancreatic cancer cell lines harboring the 8p amplicon [10,37][10][33]. 8][8].3.1.2. Fusion Proteins
Rearrangements involving the short arm of chromosome 8 have been reported and associated with different types of cancer. The first NSD3 fusion protein was found in a patient with AML, where the t(8;11) (p11.2;p15) translocation fuses the NUP98 gene to the 3′ end of NSD3 containing both of the PWWP, the SET, PHD and CH5CH rich domains (Figure 32A) [40][34]. The fusion transcript includes the FG repeats of NUP98, which are known to bind transcription factors, such as CREB-binding protein [40][34]. This suggests the importance of the transcriptional regulation of leukemic cells and indicates the NUP98-NSD3 fusion into a vital leukemogenesis-related oncogene. The presence of the NUP98-NSD3 fusion protein has been observed in leukemia cell lines and has also been found in B-lymphocyte cell lines derived from healthy volunteers who had undergone transformation by the Epstein–Barr virus [41][35].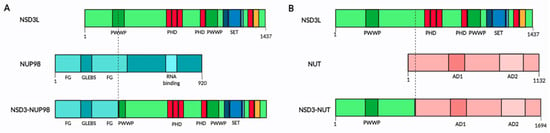
3.1.3. Mutations
Xiong et al. described a variety of NDS3 missense mutations and T419Pfs*8/Nfs*28/N mutations in four cases of stomach adenocarcinoma (STAD), two cases of colon adenocarcinoma (COAD) and single cases of breast invasive carcinoma (BRCA) and pancreatic adenocarcinoma (PAAD). Likewise, nonsense mutations in NSD3, such as, E1181K and T2342A, enhance the growth of cancer cells and xenograft tumors by disrupting an autoinhibitory loop in the NSD3 protein, thereby increasing enzymatic activity [47,48][41][42].4. NSD3 Involvement in Cancer
It is well known that NSD3 catalyzes the methylation of histone H3 at lysine 36, this occurs because NSD3 binds to LSD2 and G9a/EHMT2, forming a complex in vivo [49][43]. G9a and LSD2 mediate H3K9 methylation and H3K4 demethylation of actively transcribed genes, helping NSD3 to recognize and methylate H3K36 [14]. Morishita et al. used the C-terminal portion of NSD3, including pre-SET, SET, post-SET and PHD5 domain and identified that in vitro, NSD3 can methylate H3K9, H3K27, H3K36, H3K79 and H4K20 [4]. Discrepancies about the specificity for the substrate of the catalytic domain of NSD3 and other members of the NSD family, may be due to the cellular context, the assay employed, the nature of the substrate, or the portion of the protein used, if it is only the SET domain or the full-length protein. In relation to the isoforms of NSD3, NSD3L has been associated with neural crest formation and migration, playing a role in gene expression during neural crest development [50,51][44][45]. NSD3S conserves the N-terminal PWWP domain, this domain allows the protein to bind histone H3 at methylated lysine 36 [52][46]. The WHISTLE isoform has been found to act as a transcriptional repressor through HDAC1 recruitment, having H3K4me2 and H3K27me2/3 methyltransferase activity [25[24][47],53], this isoform is considered less relevant to cancer.4.1. Methyltransferase-Dependent Function of NSD3 in Cancer
4.1.1. NOTCH Pathway
NSD3L interacts with EZH2 and RNA polymerase II to influence H3K36me2/3-dependent transactivation of genes, including those related to NOTCH signaling in breast cancer with the 8p11-12 amplicon, such as NOTCH receptors, ligands, and ADAM12 [54][48]. These findings indicate that NSD3-induced methylation of H3K36 activates NOTCH signaling to drive breast tumor initiation and metastatic progression.4.1.2. mTOR Pathway
Deregulation of the mTOR pathway occurs in various diseases, including cancer. The mTOR pathway responds to environmental signals, regulating basic cell functions like cell growth and proliferation [55][49], survival, apoptosis, angiogenesis, and metabolism [56][50].4.1.3. EGFR Pathway
It has been shown that HMTs methylate not only histones, but also proteins. The NSD family of proteins have also been described as performing that function, as both NSD1 [57][51] and NSD2 [58][52] methylate NF-kB to regulate its function. NSD3-mediated mono-methylation of the EGFR kinase domain (Lys721) affects the cytoplasmic and nuclear function of the protein. In the cytoplasm, it increases EGFR kinase activity and the downstream ERK signaling pathway without the presence of the ligand EGF. In the nucleus, it stimulates cell cycle progression by increasing the binding of EGFR to PCNA on squamous cell carcinoma of the head and neck (SCCHN) cancer cells [59][53].4.1.4. IFN Pathway
Activated Interferon regulatory factor 3 (IRF3) is a transcriptional regulator that promotes IFN-α and IFN-β transcription. IFN-β elicits both anti-inflammatory and pro-inflammatory responses, playing a key role in innate immunity and the response to viral infections [61,62][54][55]. It is accepted that innate and adaptive immunity plays an important role in antitumor immune surveillance. Amplification of NSD3 in patients with LUSC exhibits a decrease in the type II IFN response, leading to an immune–desert pro-tumorigenic phenotype [64][56].4.1.5. Cyclin Dependent Kinase (CDK) Pathway
CDC6 and CDK2 promote G1 to S phase transitions, and the transcription of these genes is regulated by H3K36 di-methylation. In SCCHN cells, it was demonstrated that NSD3 regulates transcription of CDC6 and CDK2, as knockdown of NSD3 resulted in G0/G1 arrest [66][57]. Knockdown of NSD3 by siRNA in bladder and lung cancer cell lines reduced cell proliferation by inducing cell cycle arrest at G2/M phases through the regulation of the expression of CCNG1 and NEK7, which are important regulators of G2/M transition in cancer cells [67,68,69][58][59][60].4.2. NSD3S Isoform Function as an Adaptor Protein
4.2.1. NSD3-NUT Fusion
The NSD3-NUT fusion oncoprotein is present in several NMC cases. After knockdown of endogenous NSD3-NUT in an NMC cell line, there was an increase in keratin levels, and a decrease in cellular proliferation, indicating a crucial role of the NSD3-NUT oncofusion in blocking cell differentiation and stimulating the proliferation in this cell line. It was also found that NSD3 not only interacts with NUT, but is associated with BRD4-NUT fusion, this interaction being important in the blockade of differentiation [43][37].4.2.2. NSD3S-BRD4-CHD8 Interactions
NSD3 imparts a pTEFb-independent transcriptional activation function on BRD4, on genes such as CCND1 and PIM2. The BRD4/NSD3 complex regulates the methylation of H3K36 at BRD4 target genes [75][61]. BRD4 regulates the transcription of some genes, like CD274 that encodes PD-L1 [76][62]. NSD3S was described as an adaptor protein by Shen et al. that characterized the binding of BRD4 to a small 11 amino acid region on the N-terminal of NSD3 (amino acid 152–163). Also, they showed that the short isoform, NSD3S, was required and sufficient for driving leukemia progression, indicating a methyltransferase independent function of the protein. NSD3S also binds to the chromatin remodeler CHD8 through the C-terminal region, linking BRD4 to CHD8 on the chromatin, through the ET domain of BRD4 [23][22]. The three proteins colocalize in regions of the genome and they are release from MYC super-enhancers using BET inhibitor, JQ-1 [23][22].4.2.3. NSD3S-MYC Interaction
Sun et al. reported that in NSD3 knockout pancreatic cells and in shRNA-xenografts, there was a decrease in the gene expression of Myc, Adam12, and Notch3, demonstrating that the silencing of NSD3 can downregulate oncogenic genes [48][42]. Scholars described that NSD3S interacts with MYC to stabilize the MYC protein and increase its transcriptional activity, acting as an oncogenic interaction [77][63]. The NSD3S-MYC interaction is mediated by a 15 amino acid site on NSD3S, between amino acids 389 and 404.References
- Kornberg, R.D.; Lorch, Y. Twenty-Five Years of the Nucleosome, Fundamental Particle of the Eukaryote Chromosome. Cell 1999, 98, 285–294.
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.-Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-Resolution Profiling of Histone Methylations in the Human Genome. Cell 2007, 129, 823–837.
- Kouzarides, T. Histone Methylation in Transcriptional Control. Curr. Opin. Genet. Dev. 2002, 12, 198–209.
- Morishita, M.; Di Luccio, E. Structural Insights into the Regulation and the Recognition of Histone Marks by the SET Domain of NSD1. Biochem. Biophys. Res. Commun. 2011, 412, 214–219.
- Li, W.; Tian, W.; Yuan, G.; Deng, P.; Sengupta, D.; Cheng, Z.; Cao, Y.; Ren, J.; Qin, Y.; Zhou, Y.; et al. Molecular Basis of Nucleosomal H3K36 Methylation by NSD Methyltransferases. Nature 2021, 590, 498–503.
- Douglas, J.; Coleman, K.; Tatton-Brown, K.; Hughes, H.E.; Temple, I.K.; Cole, T.R.P.; Rahman, N. Evaluation of NSD2 and NSD3 in Overgrowth Syndromes. Eur. J. Hum. Genet. 2005, 13, 150–153.
- Tauchmann, S.; Schwaller, J. NSD1: A Lysine Methyltransferase between Developmental Disorders and Cancer. Life 2021, 11, 877.
- Toyokawa, G.; Cho, H.-S.; Masuda, K.; Yamane, Y.; Yoshimatsu, M.; Hayami, S.; Takawa, M.; Iwai, Y.; Daigo, Y.; Tsuchiya, E.; et al. Histone Lysine Methyltransferase Wolf-Hirschhorn Syndrome Candidate 1 Is Involved in Human Carcinogenesis through Regulation of the Wnt Pathway. Neoplasia 2011, 13, 887-IN11.
- Liu, L.; Kimball, S.; Liu, H.; Holowatyj, A.; Yang, Z.-Q. Genetic Alterations of Histone Lysine Methyltransferases and Their Significance in Breast Cancer. Oncotarget 2015, 6, 2466–2482.
- Mahmood, S.F.; Gruel, N.; Nicolle, R.; Chapeaublanc, E.; Delattre, O.; Radvanyi, F.; Bernard-Pierrot, I. PPAPDC1B and WHSC1L1 Are Common Drivers of the 8p11-12 Amplicon, Not Only in Breast Tumors But Also in Pancreatic Adenocarcinomas and Lung Tumors. Am. J. Pathol. 2013, 183, 1634–1644.
- Rooney, C.; Geh, C.; Williams, V.; Heuckmann, J.M.; Menon, R.; Schneider, P.; Al-Kadhimi, K.; Dymond, M.; Smith, N.R.; Baker, D.; et al. Characterization of FGFR1 Locus in sqNSCLC Reveals a Broad and Heterogeneous Amplicon. PLoS ONE 2016, 11, e0149628.
- Stec, I.; Van Ommen, G.-J.B.; Den Dunnen, J.T. WHSC1L1, on Human Chromosome 8p11.2, Closely Resembles WHSC1 and Maps to a Duplicated Region Shared with 4p16.3. Genomics 2001, 76, 5–8.
- Angrand, P.-O.; Apiou, F.; Stewart, A.F.; Dutrillaux, B.; Losson, R.; Chambon, P. NSD3, a New SET Domain-Containing Gene, Maps to 8p12 and Is Amplified in Human Breast Cancer Cell Lines. Genomics 2001, 74, 79–88.
- He, C.; Li, F.; Zhang, J.; Wu, J.; Shi, Y. The Methyltransferase NSD3 Has Chromatin-Binding Motifs, PHD5-C5HCH, That Are Distinct from Other NSD (Nuclear Receptor SET Domain) Family Members in Their Histone H3 Recognition. J. Biol. Chem. 2013, 288, 4692–4703.
- Wu, H.; Zeng, H.; Lam, R.; Tempel, W.; Amaya, M.F.; Xu, C.; Dombrovski, L.; Qiu, W.; Wang, Y.; Min, J. Structural and Histone Binding Ability Characterizations of Human PWWP Domains. PLoS ONE 2011, 6, e18919.
- Rona, G.B.; Almeida, D.S.G.; Pinheiro, A.S.; Eleutherio, E.C.A. The PWWP Domain of the Human Oncogene WHSC1L1/NSD3 Induces a Metabolic Shift toward Fermentation. Oncotarget 2017, 8, 54068–54081.
- Vermeulen, M.; Eberl, H.C.; Matarese, F.; Marks, H.; Denissov, S.; Butter, F.; Lee, K.K.; Olsen, J.V.; Hyman, A.A.; Stunnenberg, H.G.; et al. Quantitative Interaction Proteomics and Genome-Wide Profiling of Epigenetic Histone Marks and Their Readers. Cell 2010, 142, 967–980.
- Stec, I.; Nagl, S.B.; van Ommen, G.-J.B. The PWWP Domain: A Potential Protein-Protein Interaction Domain in Nuclear Proteins Influencing Differentiation? FEBS Lett. 2000, 473, 1–5.
- Jain, K.; Fraser, C.S.; Marunde, M.R.; Parker, M.M.; Sagum, C.; Burg, J.M.; Hall, N.; Popova, I.K.; Rodriguez, K.L.; Vaidya, A.; et al. Characterization of the Plant Homeodomain (PHD) Reader Family for Their Histone Tail Interactions. Epigenetics Chromatin 2020, 13, 3.
- Morishita, M.; Mevius, D.; Di Luccio, E. In Vitro Histone Lysine Methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct. Biol. 2014, 14, 1–13.
- Yang, Z.-Q.; Liu, G.; Bollig-Fischer, A.; Giroux, C.N.; Ethier, S.P. Transforming Properties of 8p11-12 Amplified Genes in Human Breast Cancer. Cancer Res. 2010, 70, 8487–8497.
- Shen, C.; Ipsaro, J.J.; Shi, J.; Milazzo, J.P.; Wang, E.; Roe, J.-S.; Suzuki, Y.; Pappin, D.J.; Joshua-Tor, L.; Vakoc, C.R. NSD3-Short Is an Adaptor Protein That Couples BRD4 to the CHD8 Chromatin Remodeler. Mol. Cell 2015, 60, 847–859.
- Irish, J.C.; Mills, J.N.; Turner-Ivey, B.; Wilson, R.C.; Guest, S.T.; Rutkovsky, A.; Dombkowski, A.; Kappler, C.S.; Hardiman, G.; Ethier, S.P. Amplification of WHSC1L1 Regulates Expression and Estrogen-independent Activation of ERα in SUM-44 Breast Cancer Cells and Is Associated with ERα Over-expression in Breast Cancer. Mol. Oncol. 2016, 10, 850–865.
- Kim, S.M.; Kee, H.J.; Eom, G.H.; Choe, N.W.; Kim, J.Y.; Kim, Y.S.; Kim, S.K.; Kook, H.; Kook, H.; Seo, S.B. Characterization of a Novel WHSC1-Associated SET Domain Protein with H3K4 and H3K27 Methyltransferase Activity. Biochem. Biophys. Res. Commun. 2006, 345, 318–323.
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium; Aaltonen, L.A.; Abascal, F.; Abeshouse, A.; Aburatani, H.; Adams, D.J.; Agrawal, N.; Ahn, K.S.; Ahn, S.-M.; Aikata, H.; et al. Pan-Cancer Analysis of Whole Genomes. Nature 2020, 578, 82–93.
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell 2011, 144, 27–40.
- Forment, J.V.; Kaidi, A.; Jackson, S.P. Chromothripsis and Cancer: Causes and Consequences of Chromosome Shattering. Nat. Rev. Cancer 2012, 12, 663–670.
- Krupina, K.; Goginashvili, A.; Cleveland, D.W. Scrambling the Genome in Cancer: Causes and Consequences of Complex Chromosome Rearrangements. Nat. Rev. Genet. 2023.
- Voutsadakis, I.A. 8p11.23 Amplification in Breast Cancer: Molecular Characteristics, Prognosis and Targeted Therapy. J. Clin. Med. 2020, 9, 3079.
- Bernard-Pierrot, I.; Gruel, N.; Stransky, N.; Vincent-Salomon, A.; Reyal, F.; Raynal, V.; Vallot, C.; Pierron, G.; Radvanyi, F.; Delattre, O. Characterization of the Recurrent 8p11-12 Amplicon Identifies PPAPDC1B, a Phosphatase Protein, as a New Therapeutic Target in Breast Cancer. Cancer Res. 2008, 68, 7165–7175.
- Turner-Ivey, B.; Smith, E.L.; Rutkovsky, A.C.; Spruill, L.S.; Mills, J.N.; Ethier, S.P. Development of Mammary Hyperplasia, Dysplasia, and Invasive Ductal Carcinoma in Transgenic Mice Expressing the 8p11 Amplicon Oncogene NSD3. Breast Cancer Res. Treat. 2017, 164, 349–358.
- Satpathy, S.; Krug, K.; Jean Beltran, P.M.; Savage, S.R.; Petralia, F.; Kumar-Sinha, C.; Dou, Y.; Reva, B.; Kane, M.H.; Avanessian, S.C.; et al. A Proteogenomic Portrait of Lung Squamous Cell Carcinoma. Cell 2021, 184, 4348–4371.e40.
- Tonon, G.; Wong, K.-K.; Maulik, G.; Brennan, C.; Feng, B.; Zhang, Y.; Khatry, D.B.; Protopopov, A.; You, M.J.; Aguirre, A.J.; et al. High-Resolution Genomic Profiles of Human Lung Cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 9625–9630.
- Chen, Y.; McGee, J.; Chen, X.; Doman, T.N.; Gong, X.; Zhang, Y.; Hamm, N.; Ma, X.; Higgs, R.E.; Bhagwat, S.V.; et al. Identification of Druggable Cancer Driver Genes Amplified across TCGA Datasets. PLoS ONE 2014, 9, e98293.
- Rosati, R.; La Starza, R.; Veronese, A.; Aventin, A.; Schwienbacher, C.; Vallespi, T.; Negrini, M.; Martelli, M.F.; Mecucci, C. NUP98 Is Fused to the NSD3 Gene in Acute Myeloid Leukemia Associated with t(8;11)(P11.2;P15). Blood 2002, 99, 3857–3860.
- Taketani, T.; Taki, T.; Nakamura, H.; Taniwaki, M.; Masuda, J.; Hayashi, Y. NUP98–NSD3 Fusion Gene in Radiation-Associated Myelodysplastic Syndrome with t(8;11)(P11;P15) and Expression Pattern of NSD Family Genes. Cancer Genet. Cytogenet. 2009, 190, 108–112.
- Avenarius, M.R.; Miller, C.R.; Arnold, M.A.; Koo, S.; Roberts, R.; Hobby, M.; Grossman, T.; Moyer, Y.; Wilson, R.K.; Mardis, E.R.; et al. Genetic Characterization of Pediatric Sarcomas by Targeted RNA Sequencing. J. Mol. Diagn. 2020, 22, 1238–1245.
- French, C.A.; Rahman, S.; Walsh, E.M.; Kühnle, S.; Grayson, A.R.; Lemieux, M.E.; Grunfeld, N.; Rubin, B.P.; Antonescu, C.R.; Zhang, S.; et al. NSD3–NUT Fusion Oncoprotein in NUT Midline Carcinoma: Implications for a Novel Oncogenic Mechanism. Cancer Discov. 2014, 4, 928–941.
- Suzuki, S.; Kurabe, N.; Ohnishi, I.; Yasuda, K.; Aoshima, Y.; Naito, M.; Tanioka, F.; Sugimura, H. NSD3-NUT-Expressing Midline Carcinoma of the Lung: First Characterization of Primary Cancer Tissue. Pathol. Res. Pract. 2015, 211, 404–408.
- Agaimy, A.; Tögel, L.; Stoehr, R.; Meidenbauer, N.; Semrau, S.; Hartmann, A.; Mantsopoulos, K. NSD3-NUTM1-Rearranged Carcinoma of the Median Neck/Thyroid Bed Developing after Recent Thyroidectomy for Sclerosing Mucoepidermoid Carcinoma with Eosinophilia: Report of an Extraordinary Case. Virchows Arch. 2021, 479, 1095–1099.
- Chen, M.; Yang, J.; Lv, L.; Li, Y.; Tang, Y.; Liu, W.; Wang, W.; Jiang, L. Comprehensive Genetic Profiling of Six Pulmonary Nuclear Protein in Testis Carcinomas with a Novel Micropapillary Histological Subtype in Two Cases. Hum. Human. Pathol. 2021, 115, 56–66.
- Xiong, Q.; Zhou, Y.; Zhang, S.; Zhang, Y.; Xu, Y.; Yang, Y.; Zhou, C.; Zeng, Z.; Han, J.; Zhu, Q. NSD3, a Member of Nuclear Receptor-Binding SET Domain Family, Is a Potential Prognostic Biomarker for Pancreatic Cancer. Cancer Med. 2023, 12, 10961–10978.
- Sun, Y.; Xie, J.; Cai, S.; Wang, Q.; Feng, Z.; Li, Y.; Lu, J.; Chen, W.; Ye, Z. Elevated Expression of Nuclear Receptor-Binding SET Domain 3 Promotes Pancreatic Cancer Cell Growth. Cell Death Dis. 2021, 12, 913.
- Fang, R.; Barbera, A.J.; Xu, Y.; Rutenberg, M.; Leonor, T.; Bi, Q.; Lan, F.; Mei, P.; Yuan, G.-C.; Lian, C.; et al. Human LSD2/KDM1b/AOF1 Regulates Gene Transcription by Modulating Intragenic H3K4me2 Methylation. Mol. Cell 2010, 39, 222–233.
- Jacques-Fricke, B.T.; Roffers-Agarwal, J.; Hussein, A.O.; Yoder, K.J.; Gearhart, M.D.; Gammill, L.S. Profiling NSD3-Dependent Neural Crest Gene Expression Reveals Known and Novel Candidate Regulatory Factors. Dev. Biol. 2021, 475, 118–130.
- Jacques-Fricke, B.T.; Gammill, L.S. Neural Crest Specification and Migration Independently Require NSD3-Related Lysine Methyltransferase Activity. MBoC 2014, 25, 4174–4186.
- Qin, S.; Min, J. Structure and Function of the Nucleosome-Binding PWWP Domain. Trends Biochem. Sci. 2014, 39, 536–547.
- Kim, S.-M.; Kee, H.-J.; Choe, N.; Kim, J.-Y.; Kook, H.; Kook, H.; Seo, S.-B. The Histone Methyltransferase Activity of WHISTLE Is Important for the Induction of Apoptosis and HDAC1-Mediated Transcriptional Repression. Exp. Cell Res. 2007, 313, 975–983.
- Jeong, G.-Y.; Park, M.K.; Choi, H.-J.; An, H.W.; Park, Y.-U.; Choi, H.-J.; Park, J.; Kim, H.-Y.; Son, T.; Lee, H.; et al. NSD3-Induced Methylation of H3K36 Activates NOTCH Signaling to Drive Breast Tumor Initiation and Metastatic Progression. Cancer Res. 2021, 81, 77–90.
- Jacinto, E.; Loewith, R.; Schmidt, A.; Lin, S.; Rüegg, M.A.; Hall, A.; Hall, M.N. Mammalian TOR Complex 2 Controls the Actin Cytoskeleton and Is Rapamycin Insensitive. Nat. Cell Biol. 2004, 6, 1122–1128.
- Thedieck, K.; Polak, P.; Kim, M.L.; Molle, K.D.; Cohen, A.; Jenö, P.; Arrieumerlou, C.; Hall, M.N. PRAS40 and PRR5-Like Protein Are New mTOR Interactors That Regulate Apoptosis. PLoS ONE 2007, 2, e1217.
- Lu, T.; Jackson, M.W.; Wang, B.; Yang, M.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-κB by NSD1/FBXL11-Dependent Reversible Lysine Methylation of P65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51.
- Yang, P.; Guo, L.; Duan, Z.J.; Tepper, C.G.; Xue, L.; Chen, X.; Kung, H.-J.; Gao, A.C.; Zou, J.X.; Chen, H.-W. Histone Methyltransferase NSD2/MMSET Mediates Constitutive NF-κB Signaling for Cancer Cell Proliferation, Survival, and Tumor Growth via a Feed-Forward Loop. Mol. Cell. Biol. 2012, 32, 3121–3131.
- Saloura, V.; Vougiouklakis, T.; Zewde, M.; Deng, X.; Kiyotani, K.; Park, J.-H.; Matsuo, Y.; Lingen, M.; Suzuki, T.; Dohmae, N.; et al. WHSC1L1-Mediated EGFR Mono-Methylation Enhances the Cytoplasmic and Nuclear Oncogenic Activity of EGFR in Head and Neck Cancer. Sci. Rep. 2017, 7, 40664.
- Lin, R.; Heylbroeck, C.; Pitha, P.M.; Hiscott, J. Virus-Dependent Phosphorylation of the IRF-3 Transcription Factor Regulates Nuclear Translocation, Transactivation Potential, and Proteasome-Mediated Degradation. Mol. Cell. Biol. 1998, 18, 2986–2996.
- Sato, M.; Hata, N.; Asagiri, M.; Nakaya, T.; Taniguchi, T.; Tanaka, N. Positive Feedback Regulation of Type I IFN Genes by the IFN-inducible Transcription Factor IRF-7. FEBS Lett. 1998, 441, 106–110.
- Xu, D.; Liu, S.; Wu, X.; Marti, T.M.; Dorn, P.; Schmid, R.A.; Peng, R.-W.; Shu, Y. Dissecting the Immunological Profiles in NSD3-Amplified LUSC through Integrative Multi-Scale Analyses. Cancers 2022, 14, 4997.
- Saloura, V.; Vougiouklakis, T.; Zewde, M.; Kiyotani, K.; Park, J.-H.; Gao, G.; Karrison, T.; Lingen, M.; Nakamura, Y.; Hamamoto, R. WHSC1L1 Drives Cell Cycle Progression through Transcriptional Regulation of CDC6 and CDK2 in Squamous Cell Carcinoma of the Head and Neck. Oncotarget 2016, 7, 42527–42538.
- Kang, D.; Cho, H.; Toyokawa, G.; Kogure, M.; Yamane, Y.; Iwai, Y.; Hayami, S.; Tsunoda, T.; Field, H.I.; Matsuda, K.; et al. The Histone Methyltransferase Wolf–Hirschhorn Syndrome Candidate 1-like 1 (WHSC1L1) Is Involved in Human Carcinogenesis. Genes. Chromosomes Cancer 2013, 52, 126–139.
- Seo, H.R.; Lee, D.H.; Lee, H.J.; Baek, M.; Bae, S.; Soh, J.W.; Lee, S.J.; Kim, J.; Lee, Y.S. Cyclin G1 Overcomes Radiation-Induced G2 Arrest and Increases Cell Death through Transcriptional Activation of Cyclin B1. Cell Death Differ. 2006, 13, 1475–1484.
- Salem, H.; Rachmin, I.; Yissachar, N.; Cohen, S.; Amiel, A.; Haffner, R.; Lavi, L.; Motro, B. Nek7 Kinase Targeting Leads to Early Mortality, Cytokinesis Disturbance and Polyploidy. Oncogene 2010, 29, 4046–4057.
- Rahman, S.; Sowa, M.E.; Ottinger, M.; Smith, J.A.; Shi, Y.; Harper, J.W.; Howley, P.M. The Brd4 Extraterminal Domain Confers Transcription Activation Independent of pTEFb by Recruiting Multiple Proteins, Including NSD3. Mol. Cell. Biol. 2011, 31, 2641–2652.
- Zhu, H.; Bengsch, F.; Svoronos, N.; Rutkowski, M.R.; Bitler, B.G.; Allegrezza, M.J.; Yokoyama, Y.; Kossenkov, A.V.; Bradner, J.E.; Conejo-Garcia, J.R.; et al. BET Bromodomain Inhibition Promotes Anti-Tumor Immunity by Suppressing PD-L1 Expression. Cell Rep. 2016, 16, 2829–2837.
- Li, Z.; Ivanov, A.A.; Su, R.; Gonzalez-Pecchi, V.; Qi, Q.; Liu, S.; Webber, P.; McMillan, E.; Rusnak, L.; Pham, C.; et al. The OncoPPi Network of Cancer-Focused Protein–Protein Interactions to Inform Biological Insights and Therapeutic Strategies. Nat. Commun. 2017, 8, 14356.
