气候变化和高温事件频率的增加对全球作物产量构成重大威胁。为了解决这个问题,全面了解植物如何应对热冲击(Climate change and the increasing frequency of high temperature (HT) events are significant threats to global crop yields. To address this, a comprehensive understanding of how plants respond to heat shock (HS)至关重要。涉及钙(S) is essential. Signaling pathways involving calcium (Ca2+),是植物中通用的第二信使,通过离子浓度的时间和空间变化来编码信息。钙), a versatile second messenger in plants, encode information through temporal and spatial variations in ion concentration. Ca2+被 is detected by Ca 检测到2+-感应效应子,包括通道和结合蛋白,触发特定的细胞反应。在高温下,sensing effectors, including channels and binding proteins, which trigger specific cellular responses. At elevated temperatures, the cytosolic concentration of Ca 的胞质浓度2+在植物细胞中迅速增加,使 in plant cells increases rapidly, making Ca2+表示对 signals the earliest response to HS 的最早反应。.
- crop yield
- thermotolerance
- signaling
- Ca2+ Signaling and Plant Thermotolerance
The rapid rise in CO2 levels on Earth is causing global warming, leading to more frequent extreme high-temperature (HT) events. Heat is a major factor in reducing crop yields. It disrupts homeostasis, affects seed germination, and alters (and ultimately stunts) plant growth [1]. Without sufficient adaptation, genetic enhancements, and fertilization, every one degree Celsius increase in global mean temperature could lead to significant reductions in global wheat, rice, and maize yields (averaging 6.0%, 3.2%, and 7.4%, respectively) [2]. Therefore, understanding the molecular mechanisms underlying plant responses to HT stress is vital for improving agricultural production and ensuring future food security.
HT can elevate the intracellular concentration of calcium (Ca2+), a common second messenger in both animal and plant cells. Plants exposed to HT can experience heat shock (HS), which triggers a rise in cytosolic Ca2+ and disrupts the oscillations in Ca2+ levels [3]. Ca2+ plays a critical role in maintaining the normal physiological functions of plant cells and is involved in various physiological processes in plants. Additionally, as a ubiquitous second messenger, Ca2+ participates in plant responses to various stressors. Thus, understanding the impact of Ca2+ on plant thermotolerance is essential for the development of heat-resistant crops [4, 5].
Ca2+ is a versatile intracellular signal; information is encoded based on temporal and spatial patterns of Ca2+ concentration changes. These patterns are decoded by Ca2+-sensing effectors such as Ca2+-permeable channels and Ca2+-binding proteins to initiate specific cellular responses [5]. The induction of the Ca2+ signal represents the most rapid response to elevated temperatures in plants. In one study, the cytosolic Ca2+ concentration in wheat peaked within 10–15 min of a sudden temperature increase from 24 to 36 °C before returning gradually to baseline as the HS response (HSR) continued [6].
Ca2+ entry into the cytoplasm is facilitated by several families of protein channels, including cyclic nucleotide-gated channels (CNGCs), glutamate (Glu) receptor-like channels (GLRs), annexins, and mechanosensitive (MS) channels. Each of these channel types plays a crucial role in promoting an influx of Ca2+. Furthermore, plants possess various Ca2+-binding proteins that decode and transmit the primary Ca2+ signal to elicit specific cellular responses. These proteins include calmodulins (CaMs), CaM-like proteins (CMLs), Ca2+-dependent protein kinases (CDPKs or CPKs), Ca2+- and CaM-dependent protein kinases (CCaMKs), calcineurin B-like proteins (CBLs), and CBL-interacting protein kinases (CIPKs). When these proteins bind Ca2+, they undergo conformational changes that allow them to initiate downstream signaling events via interactions with other proteins or molecules. This mechanism enables plants to translate variations in the intracellular Ca2+ concentrations into specific cellular responses, including changes in gene expression, enzyme activity, or ion channel activity [7-9].
Recent studies have focused on understanding how plants detect Ca2+ increases due to HS, and many studies have identified crucial molecules and signaling pathways involved. Here, we review the latest findings on thermosensing in different crop species.
- Ca2+-Permeable Channels Perceive Elevated Temperatures
Presently, the identity of a definitive thermosensor in plants remains elusive. It is hypothesized that Ca2+ channels located in the plasma membrane (PM) are crucial players in perceiving elevated temperatures. These channels could be activated directly or indirectly, leading to increased cytoplasmic Ca2+ levels. Consequently, thermosensors may function as Ca2+ channels, directly modulating Ca2+ signaling in response to external stimuli. Alternatively, they may act as regulatory elements that influence the membrane lipid composition, which is closely linked to Ca2+ channel activity. Another possibility is that they operate as GLRs, initiating an influx of Ca2+ in response to external stimuli.
Genome sequencing has revealed that plants lack typical animal Ca2+ channels such as voltage-dependent Ca2+ channels, transient receptor potential channels, purinergic P2X receptor channels, and cysteine loop channels. Instead, they have expanded families of CNGCs, GLRs, annexins, reduced hyperosmolarity-induced [Ca2+] increase channels (OSCAs), “Mid1-complementing activity” channels (MCAs), two-pore channels (TPCs), MS-like channels (MSLs), and Piezo channels (MSPs). In the context of HSR, we will explore several potential candidates among these Ca2+-permeable ion channels (Figure 1).
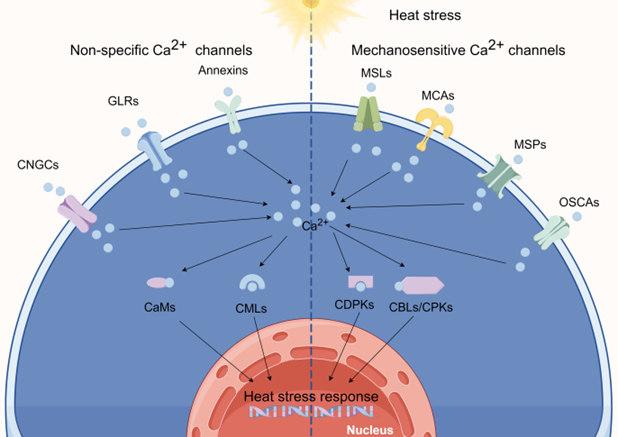
Figure 1. calcium (Ca2+) channels involved in sensing heat. (By Figdraw).
2.1. Heat Sensing via CNGCs
CNGCs are non-specific cation channels that regulate the flow of Ca2+ by binding to ligands such as cAMP and cGMP. CNGCs are activated in response to various abiotic and biotic stresses [10, 11]. Plant and animal CNGCs belong to the K-selective shaker channel family and share similarities in their amino acid sequences and overall structures [12]. They possess cyclic nucleotide-binding domains and one or more CaM-binding domains at their cytosolic N- and C-termini [13, 14].+
HS induces PM fluidization, allowing an influx of Ca2+ into the cytoplasm. Furthermore, nucleotide cyclases can elevate cAMP and cGMP levels under HS conditions, further promoting the influx of Ca2+ and activating associated signaling pathways. CNGCs were initially discovered in barley [15]; subsequent studies identified 20 and 16 CNGCs in the genomes of Arabidopsis and rice, respectively [16, 17]. Currently, plant CNGCs are known to participate in a range of biological processes [18-21].
2.2. Heat Sensing via GLRs
In mammals, ionotropic Glu receptor cation channels (iGluRs) respond to the neurotransmitter Glu, initiating Ca2+ signal cascades [22]. The first evidence of Glu signaling in plants was uncovered by Lam et al. [23], who identified the AtGLR gene family in Arabidopsis. Through sequence analysis and comparisons, homologous Glu receptor variants have been identified in dicot and monocot species [24-26]. The Arabidopsis genome contains 20 GLRs, while the rice and poplar genomes harbor 13 and 61 GLRs, respectively [27, 28]. Plant GLRs display significant sequence and structural homologies with animal iGluRs, including high degrees of amino acid sequence identity in their channel and ligand-binding domains [29].
2.3. Heat Sensing via Annexins
Annexins, an evolutionarily conserved family of proteins present in a variety of organisms, are renowned for their ability to bind to Ca2+ and phospholipids. They play pivotal roles in a plethora of cellular processes [30, 31]. Annexins exhibit peroxidase and ATPase/GTPase activities and are associated with the regulation of Ca2+ channels [32]. Their functions encompass a wide range of intracellular and extracellular phenomena, including vesicular trafficking, organization of the membrane–cytoskeleton, exocytosis, endocytosis, phagocytosis, ion channel regulation, and apoptosis [33]. The genomes of Arabidopsis, rice, and wheat contain 8, 10, and 25 annexin genes, respectively [34-36]. Annexins feature a conserved protein core domain capable of binding both Ca2+ and phospholipids alongside an N-terminal domain that varies in sequence and length among different annexins [37].
In plants, annexins participate in environmental stress responses, and they play roles in growth, development, and signaling [38]. Some plant annexins regulate the level of free cytosolic Ca2+, and certain annexins can form Ca2+-permeable channels in lipid bilayers or vesicles [39]. Moreover, plant annexins may possess peroxidase activity or ATPase/GTPase activity, each contributing to functional specificity [40]. Annexins play a broad regulatory role in diverse biochemical and cellular processes, including Ca2+ channel regulation, and in plant growth, development, and biotic and abiotic environmental stress responses [40].
2.4. Heat Sensing via OSCAs
Recently discovered MS Ca2+ channels called OSCAs have the ability to detect and respond to changes in osmotic pressure, regardless of whether it originates externally or internally. OSCAs are essential for regulating the flow of Ca2+ in plants; thus, they play a crucial role in plant growth and adaptation to environmental stress. OSCA1, the first OSCA identified in Arabidopsis thaliana, is responsible for raising cytosolic Ca2+ levels by triggering an influx of Ca2+. In fact, this channel is responsible for the increase in intracellular Ca2+ triggered by multiple stimuli in plants [41]. In total, 15, 11, 12, and 62 OSCA genes have been identified in Arabidopsis [41], rice [42], maize [43], and cotton [44], respectively.
2.5. The Functions of MSLs, MCAs, and MSPs in Plants following HT Treatment
In addition to OSCAs, plants possess several other types of MS ion channels, including MSLs, MCAs, and MSPs. These channels are permeable to Ca2+ and play crucial roles in enabling plants to respond to mechanical stimuli and changes in osmotic pressure.
MSLs are MS ion channels found in both bacteria and plants. In Arabidopsis, MSL2 and MSL3 are essential to protect plastids against bursting inside leaf epidermal cells during growth under conditions of high osmotic pressure [45]. MSL8 responds to PM distortion during pollen grain rehydration and germination [46]. Additionally, MSL10 and MSL9 exhibit MS ion channel activity in root protoplasts [47]. In rice, most OsMSL genes are expressed during reproductive growth, suggesting their involvement in plant growth, development, and stress responses [48].
While the direct involvement of these channels in the HSR has not been extensively studied, their potential role in such responses warrants further investigation.
- Ca2+-Binding Protein Involvement in the HSR
Under HS conditions, the intracellular Ca2+ concentration in plants increases, primarily due to an influx of Ca2+ from outside the cell or the release of Ca2+ from intracellular stores [49, 50]. This rise serves as a pivotal “signal” that triggers a complex signal transduction cascade. This signal, in conjunction with downstream effector proteins, including CaMs, CMLs, CBLs, and CDPKs/CPKs, serves as a molecular code that must be interpreted by the cell. Together, these proteins create a sophisticated signaling network that transmits stress signals and orchestrates a multitude of responses in plants [51]. In this section, we will concentrate on candidate Ca2+-binding proteins that were reported to be involved in the HSR in crops.
3.1. CaMs in HS Signaling
CaMs, highly conserved sensor proteins containing EF-hand motifs, are found in various plant organelles. In total, 9, 5, and 8 CaM genes have been identified in Arabidopsis [52, 53], rice [54], and maize [55], respectively. CaMs are ubiquitously expressed in all eukaryotic cells and possess a dumbbell-shaped structure. They are multifunctional, consisting of two globular lobes at the N- and C-termini connected by a flexible central linker. Each lobe contains a pair of EF-hand motifs capable of binding Ca2+ ions with positive cooperativity [56]. The EF-hand motif in CaM is characterized by a helix–loop–helix (D-X-D) conformation, consisting of 12 amino acid residues, totaling 36 amino acid residues across both motifs. In the D-X-D motif, the 14th and 16th positions are fixed, while the 15th position can be occupied by any amino acid [57]. These motifs are involved in interactions with a diverse array of downstream target proteins, including ion channels, pumps, antiporters, kinases, phosphatases, transcription factors, and enzymes involved in metabolic pathways. Arabidopsis CaM isoforms CaM1/4, CaM2/3/5, CaM6, and CaM7 were found to bind to CNGC6 to varying degrees, and this binding was dependent on the presence of Ca2+ and IQ6, an atypical isoleucine–glutamine motif in CNGC6. Knockout of CaM2, CaM3, CaM5, and CaM7 genes led to a marked increase in PM inward Ca2+ current under HS conditions; however, knockout of CaM1, CaM4, and CaM6 genes had no significant effect on PM Ca2+ current [58].In some instances, these motifs function as transcription factors, recognizing and binding to target genes, thereby modulating stress responses in plants.
3.2. CMLs in HS Signaling
CMLs differ from CaMs in their length and the number of EF-hand motifs they possess. While CaMs typically contain four EF-hand motifs, CMLs can have varying numbers, ranging from one to six. CMLs are termed “CaM-like” due to their 15% amino acid identity with CaM. These proteins play a crucial role in physiological responses to various stresses, including salinity, drought, heat, and cold. In Arabidopsis, 50 CMLs have been identified, compared with 32 in rice [59].
3.3. CDPKs in HS Signaling
CDPKs, or CPKs, belong to a class of protein kinases that possess four C-terminal EF-hand motifs, which enable them to sense changes in intracellular Ca2+ levels. CDPKs are effector proteins that play pivotal roles in regulating a wide range of physiological processes, including environmental stress responses, in various plant cell types [60]. CDPKs consist of several domains, including a variable N-terminal domain, a serine/threonine protein kinase domain, an autoinhibitory junction domain (JD), and a C-terminal CML regulatory domain (CaMLD) connected by a tether. The CaMLD is composed of four EF-hand Ca2+-binding motifs. At low Ca2+ concentrations, the C-lobe of a CDPK already contains Ca2+ and interacts with the JD, stabilizing its conformation. The JD forms a helical structure that blocks substrate access by being buried within the active site of the kinase domain due to an intramolecular interaction. When the concentration of Ca2+ rises, both the N-lobe and C-lobe of the CaMLD interact with the JD, leading to a substantial conformational change that releases the active site [61].
The CDPK family is diverse, with multiple members and isoforms across plant species. Recent whole-genome expression analyses have shed light on the transcriptional regulation of CDPKs in response to various stresses, including HS, in important crop species [62-67]. These findings provide valuable insight into the functional roles of CDPKs and their potential utility in enhancing the thermotolerance of crops.
3.4. CBLs and CIPKs in HS Signaling
CBLs comprise a distinct family of Ca2+ sensors in plants with essential roles in Ca2+ signaling pathways. Specifically, they interact with and modulate the activity of CIPKs. CBLs bear resemblance to the calcineurin B-subunit in yeast and neuronal Ca2+ sensors in animals [68]. CBLs possess four EF-hands, which are Ca2+-binding motifs found in various proteins. EF-hands are structural domains characterized by a D-X-D structure, with the loop region serving as the Ca2+-binding site. These domains allow proteins to function as Ca2+ sensors that can respond to changing levels of intracellular Ca2+. In the case of CBLs, the four EF-hand domains facilitate the capture of Ca2+ ions, enabling them to play pivotal roles in numerous plant physiological processes [69]. Notably, CBLs lack intrinsic kinase activity. To transmit signals, they must form complexes with CIPKs, which are serine/threonine kinases found in plants. CIPKs possess a functional kinase domain but remain in an inactive state because of the autoinhibition caused by an interaction between the kinase domain and regulatory domain. An inhibitory motif known as NAF/FISL blocks the active site in CIPKs, preventing substrate binding and subsequent phosphorylation. This autoinhibitory mechanism keeps CIPKs in an inactive state until they are activated by binding to a CBL. Once active, the CIPK can regulate downstream proteins. This activation process is crucial for the proper functioning of the CBL–CIPK signaling pathway in plants [70]. When plants encounter stress, such as HS, the intracellular concentration of Ca2+ rises. This increase enables Ca2+ ions to bind to the EF-hand motifs in CBLs. This binding promotes the interaction of CBLs with the NAF/FISL element in CIPKs. Consequently, the CIPKs become active and can participate in stress response pathways by phosphorylating downstream proteins.
- Ca2+ Signaling Networks Mediate the Plant HSR
Ca2+ are ubiquitous second messengers in eukaryotes, participating in a wide array of signaling pathways and responses to various environmental conditions. As mentioned above, when plants experience HS, the cytosolic Ca2+ concentration rises because of an influx of Ca2+ facilitated by membrane-localized Ca2+ permeable cation channels. These ions subsequently bind to Ca2+-binding proteins, initiating signal transmission to their respective downstream pathways. In recent years, substantial progress has been made in understanding the mechanisms related to thermotolerance in plants.
The HSR signaling pathways in plants include the Ca2+ dependent, ROS, NO, HSF-HSP, HSF-independent, hydrogen sulfide (H2S), and unfolded protein response (UPR) pathways, etc. [4, 71]. There are interactions and crossovers between different HS pathways. For example, the Ca2+ signaling may intersect with the ROS, NO, and HSF-HSP pathways, forming a complex signaling network [4]. Additionally, in the HSF-independent pathway, Ca2+ might contribute to the activation of certain transcription factors independent of HSF [72, 73]. The H2S pathway introduces a novel dimension, suggesting that Ca2+ may modulate signaling events in conjunction with H2S [74, 75]. The association of Ca2+ with endoplasmic reticulum (ER) stress responses suggests a potential link between Ca2+ and UPR [76, 77]. These propositions underscore the versatility of Ca2+ signaling and its potential contributions to diverse HSR pathways in plants. Future research is needed to experimentally validate these hypotheses and enhance our understanding of the intricate molecular mechanisms governing plant responses to HS.
In this section, we consider several potential downstream pathways of Ca2+ in response to HS in plants (Figure 2).
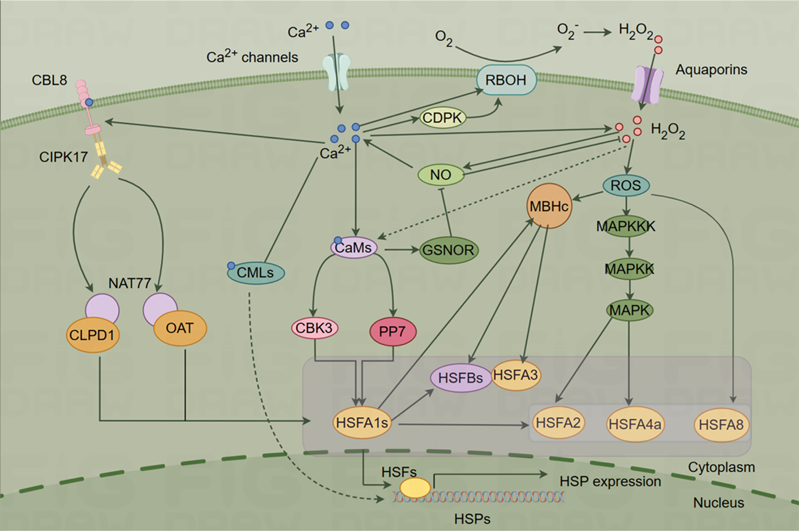
Figure 2. Ca2+ signaling pathway under heat shock (HS) (By Figdraw).
- Conclusions and Perspectives
In addition, the complex interplay of signaling pathways, including Ca2+, NO, ROS, and others, deserves further investigation. Based on the above-mentioned research gaps, future research should prioritize the following areas: (1) Integration of existing pathways. There is a need to integrate various fragmented pathways into a unified primary Ca2+ signaling pathway associated with HS. This endeavor could result in a more comprehensive understanding of the role of Ca2+ signaling in heat resistance. (2) Identification of new components. Ongoing efforts should focus on identifying new components that sense Ca2+ signals induced by HS. This continuing exploration will contribute to an improved understanding of the HSR pathway in plants. (3) Ca2+ crosstalk. Understanding the crosstalk between Ca2+-mediated HSR and other stress-signaling pathways is essential. This knowledge can shed light on the broader regulatory network that governs plant responses to multiple stressors.
A deeper understanding of the molecular mechanisms may enable the identification of key genes and pathways so as to provide targets for genetic engineering. Moreover, marker-assisted selection, next-generation molecular breeding, precision breeding, and genome editing techniques represent powerful tools to enhance the efficiency of plant breeding programs. These methods will allow breeders to select plants with desirable traits more accurately and quickly and speed up the development of crops better suited to elevated temperatures. These improvements will be conducive to promoting sustainable agriculture.
- Fitter, A. H.; Fitter, R. S. R., Rapid changes in flowering time in british plants. Science. 2002, 296, (5573), 1689-91.
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D. B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; Durand, J.-L.; Elliott, J.; Ewert, F.; Janssens, I. A.; Li, T.; Lin, E.; Liu, Q.; Martre, P.; Muller, C.; Peng, S.; Penuelas, J.; Ruane, A. C.; Wallach, D.; Wang, T.; Wu, D.; Liu, Z.; Zhu, Y.; Zhu, Z.; Asseng, S., Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. 2017, 114, (35), 9326-9331.
- Weigand, C.; Kim, S. H.; Brown, E.; Medina, E.; Mares, M.; Miller, G.; Harper, J. F.; Choi, W. G., A ratiometric calcium reporter cgf reveals calcium dynamics both in the single cell and whole plant levels under heat stress. Front. Plant Sci. 2021, 12, 777975.
- Li, B.; Gao, K.; Ren, H.; Tang, W., Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, (9), 757-779.
- Dodd, A. N.; Kudla, J.; Sanders, D., The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593-620.
- Liu, H.-T.; Li, B.; Shang, Z.-L.; Li, X.-Z.; Mu, R.-L.; Sun, D.-Y.; Zhou, R.-G., Calmodulin is involved in heat shock signal transduction in wheat. Plant physiology. 2003, 132, (3), 1186-95.
- Batistič, O.; Kudla, J., Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta. 2012, 1820, (8), 1283-93.
- Pirayesh, N.; Giridhar, M.; Ben Khedher, A.; Vothknecht, U. C.; Chigri, F., Organellar calcium signaling in plants: An update. Biochim. Biophys. Acta. Mol. Cell Res. 2021, 1868, (4), 118948.
- Kudla, J.; Batistic, O.; Hashimoto, K., Calcium signals: The lead currency of plant information processing. Plant Cell. 2010, 22, (3), 541-563.
- Swarbreck, S. M.; Colaço, R.; Davies, J. M., Plant calcium-permeable channels. Plant Physiol. 2013, 163, (2), 514-22.
- Jammes, F.; Hu, H. C.; Villiers, F.; Bouten, R.; Kwak, J. M., Calcium-permeable channels in plant cells. Febs j. 2011, 278, (22), 4262-76.
- Kaplan, B.; Sherman, T.; Fromm, H., Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007, 581, (12), 2237-46.
- DeFalco, T. A.; Marshall, C. B.; Munro, K.; Kang, H. G.; Moeder, W.; Ikura, M.; Snedden, W. A.; Yoshioka, K., Multiple calmodulin-binding sites positively and negatively regulate arabidopsis cyclic nucleotide-gated channel 12. Plant Cell. 2016, 28, (7), 1738-51.
- Fischer, C.; Kugler, A.; Hoth, S.; Dietrich, P., An iq domain mediates the interaction with calmodulin in a plant cyclic nucleotide-gated channel. Plant Cell Physiol. 2013, 54, (4), 573-84.
- Schuurink, R. C.; Shartzer, S. F.; Fath, A.; Jones, R. L., Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. 1998, 95, (4), 1944-1949.
- Mäser, P.; Thomine, S.; Schroeder, J. I.; Ward, J. M.; Hirschi, K.; Sze, H.; Talke, I. N.; Amtmann, A.; Maathuis, F. J.; Sanders, D.; Harper, J. F.; Tchieu, J.; Gribskov, M.; Persans, M. W.; Salt, D. E.; Kim, S. A.; Guerinot, M. L., Phylogenetic relationships within cation transporter families of arabidopsis. Plant Physiol. 2001, 126, (4), 1646-67.
- Bridges, D.; Fraser, M. E.; Moorhead, G. B., Cyclic nucleotide binding proteins in the arabidopsis thaliana and oryza sativa genomes. BMC Bioinform. 2005, 6, 6.
- Zhou, L.; Lan, W.; Jiang, Y.; Fang, W.; Luan, S., A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol. Plant. 2014, 7, (2), 369-76.
- Zhang, S.; Pan, Y.; Tian, W.; Dong, M.; Zhu, H.; Luan, S.; Li, L., Arabidopsis cngc14 mediates calcium influx required for tipgrowth in root hairs. Mol. Plant. 2017, 10, (7), 1004-1006.
- Chiasson, D. M.; Haage, K.; Sollweck, K.; Brachmann, A.; Dietrich, P.; Parniske, M., A quantitative hypermorphic cngc allele confers ectopic calcium flux and impairs cellular development. Elife. 2017, 6, e25012.
- Wang, Y.; Kang, Y.; Ma, C.; Miao, R.; Wu, C.; Long, Y.; Ge, T.; Wu, Z.; Hou, X.; Zhang, J.; Qi, Z., Cngc2 is a ca2+ influx channel that prevents accumulation of apoplastic ca2+ in the leaf. Plant Physiol. 2017, 173, (2), 1342-1354.
- Mayer, M. L., Structural biology of glutamate receptor ion channel complexes. Curr. Opin. Struct. Biol. 2016, 41, 119-127.
- Lam, H. M.; Chiu, J.; Hsieh, M. H.; Meisel, L.; Oliveira, I. C.; Shin, M.; Coruzzi, G., Glutamate-receptor genes in plants. Nature. 1998, 396, (6707), 125-126.
- Lacombe, B.; Becker, D.; Hedrich, R.; DeSalle, R.; Hollmann, M.; Kwak, J. M.; Schroeder, J. I.; Le Novère, N.; Nam, H. G.; Spalding, E. P.; Tester, M.; Turano, F. J.; Chiu, J.; Coruzzi, G., The identity of plant glutamate receptors. Science. 2001, 292, (5521), 1486-7.
- Li, J.; Zhu, S.; Song, X.; Shen, Y.; Chen, H.; Yu, J.; Yi, K.; Liu, Y.; Karplus, V. J.; Wu, P.; Deng, X. W., A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell. 2006, 18, (2), 340-9.
- Dietrich, P.; Anschütz, U.; Kugler, A.; Becker, D., Physiology and biophysics of plant ligand-gated ion channels. Plant Biol. 2010, 12 Suppl 1, 80-93.
- Roy, B. C.; Mukherjee, A., Computational analysis of the glutamate receptor gene family of arabidopsis thaliana. J. Biomol. Struct. Dyn. 2017, 35, (11), 2454-2474.
- Ward, J. M.; Mäser, P.; Schroeder, J. I., Plant ion channels: Gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 2009, 71, 59-82.
- Davenport, R., Glutamate receptors in plants. Ann. Bot. 2002, 90, (5), 549-57.
- Mortimer, J. C.; Laohavisit, A.; Macpherson, N.; Webb, A.; Brownlee, C.; Battey, N. H.; Davies, J. M., Annexins: Multifunctional components of growth and adaptation. J. Exp. Bot. 2008, 59, (3), 533-44.
- Davies, J. M., Annexin-mediated calcium signalling in plants. Plants-Basel. 2014, 3, (1), 128-40.
- Gorecka, K. M.; Konopka-Postupolska, D.; Hennig, J.; Buchet, R.; Pikula, S., Peroxidase activity of annexin 1 from arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2005, 336, (3), 868-75.
- Gerke, V.; Creutz, C. E.; Moss, S. E., Annexins: Linking ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, (6), 449-61.
- Clark, G. B.; Sessions, A.; Eastburn, D. J.; Roux, S. J., Differential expression of members of the annexin multigene family in arabidopsis. Plant Physiol. 2001, 126, (3), 1072-84.
- Jami, S. K.; Clark, G. B.; Ayele, B. T.; Roux, S. J.; Kirti, P. B., Identification and characterization of annexin gene family in rice. Plant Cell Rep. 2012, 31, (5), 813-25.
- Xu, L.; Tang, Y.; Gao, S.; Su, S.; Hong, L.; Wang, W.; Fang, Z.; Li, X.; Ma, J.; Quan, W.; Sun, H.; Li, X.; Wang, Y.; Liao, X.; Gao, J.; Zhang, F.; Li, L.; Zhao, C., Comprehensive analyses of the annexin gene family in wheat. BMC Genom. 2016, 17, 415.
- Gerke, V.; Moss, S. E., Annexins: From structure to function. Physiol. Rev. 2002, 82, (2), 331-71.
- Saad, R. B.; Ben Romdhane, W.; Ben Hsouna, A.; Mihoubi, W.; Harbaoui, M.; Brini, F., Insights into plant annexins function in abiotic and biotic stress tolerance. Plant Signal Behav. 2020, 15, (1), 1699264.
- Laohavisit, A.; Davies, J. M., Annexins. New Phytol. 2011, 189, (1), 40-53.
- Wu, X.; Wang, Y.; Bian, Y.; Ren, Y.; Xu, X.; Zhou, F.; Ding, H., A critical review on plant annexin: Structure, function, and mechanism. Plant Physiol. Biochem. 2022, 190, 81-89.
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; Johnson, D. M.; Swift, G. B.; He, Y.; Siedow, J. N.; Pei, Z. M., Osca1 mediates osmotic-stress-evoked ca2+ increases vital for osmosensing in arabidopsis. Nature. 2014, 514, (7522), 367-71.
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; Pei, Z. M.; Han, S., Genome-wide survey and expression analysis of the osca gene family in rice. BMC Plant Biol. 2015, 15, 261.
- Ding, S.; Feng, X.; Du, H.; Wang, H., Genome-wide analysis of maize osca family members and their involvement in drought stress. PeerJ. 2019, 7, e6765.
- Yang, X.; Xu, Y.; Yang, F.; Magwanga, R. O.; Cai, X.; Wang, X.; Wang, Y.; Hou, Y.; Wang, K.; Liu, F.; Zhou, Z., Genome-wide identification of osca gene family and their potential function in the regulation of dehydration and salt stress in gossypium hirsutum. J. Cotton Res. 2019, 2, (1), 11.
- Haswell, E. S.; Meyerowitz, E. M., Mscs-like proteins control plastid size and shape in arabidopsis thaliana. Curr. Biol. 2006, 16, (1), 1-11.
- Hamilton, E. S.; Haswell, E. S., The tension-sensitive ion transport activity of msl8 is critical for its function in pollen hydration and germination. Plant Cell Physiol. 2017, 58, (7), 1222-1237.
- Peyronnet, R.; Haswell, E. S.; Barbier-Brygoo, H.; Frachisse, J. M., Atmsl9 and atmsl10: Sensors of plasma membrane tension in arabidopsis roots. Plant Signal. Behav. 2008, 3, (9), 726-9.
- Li, J.; Zhang, M.; Sun, J.; Mao, X.; Wang, J.; Wang, J.; Liu, H.; Zheng, H.; Zhen, Z.; Zhao, H.; Zou, D., Genome-wide characterization and identification of trihelix transcription factor and expression profiling in response to abiotic stresses in rice (oryza sativa l.). Int. J. Mol. Sci. 2019, 20, (2), 251.
- Finka, A.; Cuendet, A. F.; Maathuis, F. J.; Saidi, Y.; Goloubinoff, P., Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell. 2012, 24, (8), 3333-48.
- Knight, H., Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 2000, 195, 269-324.
- Mohanta, T. K.; Yadav, D.; Khan, A. L.; Hashem, A.; Abd Allah, E. F.; Al-Harrasi, A., Molecular players of ef-hand containing calcium signaling event in plants. Int. J. Mol. Sci. 2019, 20, (6), 1476.
- Braam, J.; Davis, R. W., Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in arabidopsis. Cell. 1990, 60, (3), 357-364.
- Al-Quraan, N. A.; Locy, R. D.; Singh, N. K., Expression of calmodulin genes in wild type and calmodulin mutants of arabidopsis thaliana under heat stress. Plant Physiol. Biochem. 2010, 48, (8), 697-702.
- Choi, M. S.; Kim, M. C.; Yoo, J. H.; Moon, B. C.; Koo, S. C.; Park, B. O.; Lee, J. H.; Koo, Y. D.; Han, H. J.; Lee, S. Y.; Chung, W. S.; Lim, C. O.; Cho, M. J., Isolation of a calmodulin-binding transcription factor from rice (oryza sativa l.). J. Biol. Chem. 2005, 280, (49), 40820-31.
- Hardoim, P. R.; de Carvalho, T. L.; Ballesteros, H. G.; Bellieny-Rabelo, D.; Rojas, C. A.; Venancio, T. M.; Ferreira, P. C.; Hemerly, A. S., Genome-wide transcriptome profiling provides insights into the responses of maize (zea mays l.) to diazotrophic bacteria. Plant Soil. 2020, 451, 121-143.
- Gifford, J. L.; Walsh, M. P.; Vogel, H. J., Structures and metal-ion-binding properties of the ca2+-binding helix-loop-helix ef-hand motifs. Biochem. J. 2007, 405, (2), 199-221.
- Kundu, P.; Nehra, A.; Gill, R.; Tuteja, N.; Gill, S. S., Unraveling the importance of ef-hand-mediated calcium signaling in plants. S. Afr. J. Bot. 2022, 148, 615-633.
- Niu, W.-T.; Han, X.-W.; Wei, S.-S.; Shang, Z.-L.; Wang, J.; Yang, D.-W.; Fan, X.; Gao, F.; Zheng, S.-Z.; Bai, J.-T.; Zhang, B.; Wang, Z.-X.; Li, B.; Napier, R., Arabidopsis cyclic nucleotide-gated channel 6 is negatively modulated by multiple calmodulin isoforms during heat shock. J. Exp. Bot. 2020, 71, (1), 90-104.
- McCormack, E.; Tsai, Y. C.; Braam, J., Handling calcium signaling: Arabidopsis cams and cmls. Trends Plant Sci. 2005, 10, (8), 383-9.
- Atif, R. M.; Shahid, L.; Waqas, M.; Ali, B.; Rashid, M. A. R.; Azeem, F.; Nawaz, M. A.; Wani, S. H.; Chung, G., Insights on calcium-dependent protein kinases (cpks) signaling for abiotic stress tolerance in plants. Int. J. Mol. Sci. 2019, 20, (21), 5298.
- Yip Delormel, T.; Boudsocq, M., Properties and functions of calcium-dependent protein kinases and their relatives in arabidopsis thaliana. New Phytol. 2019, 224, (2), 585-604.
- Hu, Z.; Lv, X.; Xia, X.; Zhou, J.; Shi, K.; Yu, J.; Zhou, Y., Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Sci. 2016, 7, 469.
- Zhang, K.; Han, Y. T.; Zhao, F. L.; Hu, Y.; Gao, Y. R.; Ma, Y. F.; Zheng, Y.; Wang, Y. J.; Wen, Y. Q., Genome-wide identification and expression analysis of the cdpk gene family in grape, vitis spp. BMC Plant Biol. 2015, 15, 164.
- Zhang, H.; Liu, W. Z.; Zhang, Y.; Deng, M.; Niu, F.; Yang, B.; Wang, X.; Wang, B.; Liang, W.; Deyholos, M. K.; Jiang, Y. Q., Identification, expression and interaction analyses of calcium-dependent protein kinase (cpk) genes in canola (brassica napus l.). BMC Genom. 2014, 15, 211.
- Wu, P.; Wang, W.; Duan, W.; Li, Y.; Hou, X., Comprehensive analysis of the cdpk-snrk superfamily genes in chinese cabbage and its evolutionary implications in plants. Front. Plant Sci. 2017, 8, 162.
- Xu, X.; Liu, M.; Lu, L.; He, M.; Qu, W.; Xu, Q.; Qi, X.; Chen, X., Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 2015, 290, (4), 1403-14.
- Cai, H.; Cheng, J.; Yan, Y.; Xiao, Z.; Li, J.; Mou, S.; Qiu, A.; Lai, Y.; Guan, D.; He, S., Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genes in capsicum annuum. Front. Plant Sci. 2015, 6, 737.
- Liu, J.; Zhu, J. K., A calcium sensor homolog required for plant salt tolerance. Science. 1998, 280, (5371), 1943-1945.
- Luan, S., The cbl-cipk network in plant calcium signaling. Trends Plant Sci. 2009, 14, (1), 37-42.
- Sanyal, S. K.; Mahiwal, S.; Nambiar, D. M.; Pandey, G. K., Cbl-cipk module-mediated phosphoregulation: Facts and hypothesis. Biochem. J. 2020, 477, (5), 853-871.
- He, H.; Garcia-Mata, C.; He, L.-F., Interaction between hydrogen sulfide and hormones in plant physiological responses. Plant Growth Regul. 2018, 87, (1), 175-186.
- Yang, F.; Dong, F.-s.; Hu, F.-h.; Liu, Y.-w.; Chai, J.-f.; Zhao, H.; Lv, M.-y.; Zhou, S., Genome-wide identification and expression analysis of the calmodulin-binding transcription activator (camta) gene family in wheat (triticum aestivum l.). BMC Genet. 2020, 21, (1), 105.
- Kidokoro, S.; Konoura, I.; Soma, F.; Shinozaki, K.; Suzuki, T.; Tanokura, M.; Miyakawa, T.; Imaizumi, T.; Yamaguchi-Shinozaki, K., Clock-regulated coactivators selectively control gene expression in response to different temperature stress conditions in arabidopsis. Proc. Natl. Acad. Sci. . 2023, 120, (16), e2216183120.
- Li, Z. G.; Gong, M.; Xie, H.; Yang, L.; Li, J., Hydrogen sulfide donor sodium hydrosulfide-induced heat tolerance in tobacco (nicotiana tabacum l.) suspension cultured cells and involvement of ca2+ and calmodulin. Plant Sci. 2012, 185, 185-189.
- Wang, L.; Wan, R.; Shi, Y.; Xue, S., Hydrogen sulfide activates s-type anion channel via ost1 and ca2+ modules. Mol. Plant. 2016, 9, (3), 489-491.
- Krebs, J.; Agellon, L. B.; Michalak, M., Ca2+ homeostasis and endoplasmic reticulum (er) stress: An integrated view of calcium signaling. Biochem. Biophys. Res. Commun. 2015, 460, (1), 114-121.
- Carreras-Sureda, A.; Pihán, P.; Hetz, C., Calcium signaling at the endoplasmic reticulum: Fine-tuning stress responses. Cell Calcium. 2018, 70, 24-31.
- Fitter, A. H.; Fitter, R. S. R., Rapid changes in flowering time in british plants. Science. 2002, 296, (5573), 1689-91.
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D. B.; Huang, Y.; Huang, M.; Yao, Y.; Bassu, S.; Ciais, P.; Durand, J.-L.; Elliott, J.; Ewert, F.; Janssens, I. A.; Li, T.; Lin, E.; Liu, Q.; Martre, P.; Muller, C.; Peng, S.; Penuelas, J.; Ruane, A. C.; Wallach, D.; Wang, T.; Wu, D.; Liu, Z.; Zhu, Y.; Zhu, Z.; Asseng, S., Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. 2017, 114, (35), 9326-9331.
- Weigand, C.; Kim, S. H.; Brown, E.; Medina, E.; Mares, M.; Miller, G.; Harper, J. F.; Choi, W. G., A ratiometric calcium reporter cgf reveals calcium dynamics both in the single cell and whole plant levels under heat stress. Front. Plant Sci. 2021, 12, 777975.
- Li, B.; Gao, K.; Ren, H.; Tang, W., Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, (9), 757-779.
- Dodd, A. N.; Kudla, J.; Sanders, D., The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593-620.
- Liu, H.-T.; Li, B.; Shang, Z.-L.; Li, X.-Z.; Mu, R.-L.; Sun, D.-Y.; Zhou, R.-G., Calmodulin is involved in heat shock signal transduction in wheat. Plant physiology. 2003, 132, (3), 1186-95.
- Batistič, O.; Kudla, J., Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta. 2012, 1820, (8), 1283-93.
- Pirayesh, N.; Giridhar, M.; Ben Khedher, A.; Vothknecht, U. C.; Chigri, F., Organellar calcium signaling in plants: An update. Biochim. Biophys. Acta. Mol. Cell Res. 2021, 1868, (4), 118948.
- Kudla, J.; Batistic, O.; Hashimoto, K., Calcium signals: The lead currency of plant information processing. Plant Cell. 2010, 22, (3), 541-563.
- Swarbreck, S. M.; Colaço, R.; Davies, J. M., Plant calcium-permeable channels. Plant Physiol. 2013, 163, (2), 514-22.
- Jammes, F.; Hu, H. C.; Villiers, F.; Bouten, R.; Kwak, J. M., Calcium-permeable channels in plant cells. Febs j. 2011, 278, (22), 4262-76.
- Kaplan, B.; Sherman, T.; Fromm, H., Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007, 581, (12), 2237-46.
- DeFalco, T. A.; Marshall, C. B.; Munro, K.; Kang, H. G.; Moeder, W.; Ikura, M.; Snedden, W. A.; Yoshioka, K., Multiple calmodulin-binding sites positively and negatively regulate arabidopsis cyclic nucleotide-gated channel 12. Plant Cell. 2016, 28, (7), 1738-51.
- Fischer, C.; Kugler, A.; Hoth, S.; Dietrich, P., An iq domain mediates the interaction with calmodulin in a plant cyclic nucleotide-gated channel. Plant Cell Physiol. 2013, 54, (4), 573-84.
- Schuurink, R. C.; Shartzer, S. F.; Fath, A.; Jones, R. L., Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. 1998, 95, (4), 1944-1949.
- Mäser, P.; Thomine, S.; Schroeder, J. I.; Ward, J. M.; Hirschi, K.; Sze, H.; Talke, I. N.; Amtmann, A.; Maathuis, F. J.; Sanders, D.; Harper, J. F.; Tchieu, J.; Gribskov, M.; Persans, M. W.; Salt, D. E.; Kim, S. A.; Guerinot, M. L., Phylogenetic relationships within cation transporter families of arabidopsis. Plant Physiol. 2001, 126, (4), 1646-67.
- Bridges, D.; Fraser, M. E.; Moorhead, G. B., Cyclic nucleotide binding proteins in the arabidopsis thaliana and oryza sativa genomes. BMC Bioinform. 2005, 6, 6.
- Zhou, L.; Lan, W.; Jiang, Y.; Fang, W.; Luan, S., A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol. Plant. 2014, 7, (2), 369-76.
- Zhang, S.; Pan, Y.; Tian, W.; Dong, M.; Zhu, H.; Luan, S.; Li, L., Arabidopsis cngc14 mediates calcium influx required for tipgrowth in root hairs. Mol. Plant. 2017, 10, (7), 1004-1006.
- Chiasson, D. M.; Haage, K.; Sollweck, K.; Brachmann, A.; Dietrich, P.; Parniske, M., A quantitative hypermorphic cngc allele confers ectopic calcium flux and impairs cellular development. Elife. 2017, 6, e25012.
- Wang, Y.; Kang, Y.; Ma, C.; Miao, R.; Wu, C.; Long, Y.; Ge, T.; Wu, Z.; Hou, X.; Zhang, J.; Qi, Z., Cngc2 is a ca2+ influx channel that prevents accumulation of apoplastic ca2+ in the leaf. Plant Physiol. 2017, 173, (2), 1342-1354.
- Mayer, M. L., Structural biology of glutamate receptor ion channel complexes. Curr. Opin. Struct. Biol. 2016, 41, 119-127.
- Lam, H. M.; Chiu, J.; Hsieh, M. H.; Meisel, L.; Oliveira, I. C.; Shin, M.; Coruzzi, G., Glutamate-receptor genes in plants. Nature. 1998, 396, (6707), 125-126.
- Lacombe, B.; Becker, D.; Hedrich, R.; DeSalle, R.; Hollmann, M.; Kwak, J. M.; Schroeder, J. I.; Le Novère, N.; Nam, H. G.; Spalding, E. P.; Tester, M.; Turano, F. J.; Chiu, J.; Coruzzi, G., The identity of plant glutamate receptors. Science. 2001, 292, (5521), 1486-7.
- Li, J.; Zhu, S.; Song, X.; Shen, Y.; Chen, H.; Yu, J.; Yi, K.; Liu, Y.; Karplus, V. J.; Wu, P.; Deng, X. W., A rice glutamate receptor-like gene is critical for the division and survival of individual cells in the root apical meristem. Plant Cell. 2006, 18, (2), 340-9.
- Dietrich, P.; Anschütz, U.; Kugler, A.; Becker, D., Physiology and biophysics of plant ligand-gated ion channels. Plant Biol. 2010, 12 Suppl 1, 80-93.
- Roy, B. C.; Mukherjee, A., Computational analysis of the glutamate receptor gene family of arabidopsis thaliana. J. Biomol. Struct. Dyn. 2017, 35, (11), 2454-2474.
- Ward, J. M.; Mäser, P.; Schroeder, J. I., Plant ion channels: Gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 2009, 71, 59-82.
- Davenport, R., Glutamate receptors in plants. Ann. Bot. 2002, 90, (5), 549-57.
- Mortimer, J. C.; Laohavisit, A.; Macpherson, N.; Webb, A.; Brownlee, C.; Battey, N. H.; Davies, J. M., Annexins: Multifunctional components of growth and adaptation. J. Exp. Bot. 2008, 59, (3), 533-44.
- Davies, J. M., Annexin-mediated calcium signalling in plants. Plants-Basel. 2014, 3, (1), 128-40.
- Gorecka, K. M.; Konopka-Postupolska, D.; Hennig, J.; Buchet, R.; Pikula, S., Peroxidase activity of annexin 1 from arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2005, 336, (3), 868-75.
- Gerke, V.; Creutz, C. E.; Moss, S. E., Annexins: Linking ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, (6), 449-61.
- Clark, G. B.; Sessions, A.; Eastburn, D. J.; Roux, S. J., Differential expression of members of the annexin multigene family in arabidopsis. Plant Physiol. 2001, 126, (3), 1072-84.
- Jami, S. K.; Clark, G. B.; Ayele, B. T.; Roux, S. J.; Kirti, P. B., Identification and characterization of annexin gene family in rice. Plant Cell Rep. 2012, 31, (5), 813-25.
- Xu, L.; Tang, Y.; Gao, S.; Su, S.; Hong, L.; Wang, W.; Fang, Z.; Li, X.; Ma, J.; Quan, W.; Sun, H.; Li, X.; Wang, Y.; Liao, X.; Gao, J.; Zhang, F.; Li, L.; Zhao, C., Comprehensive analyses of the annexin gene family in wheat. BMC Genom. 2016, 17, 415.
- Gerke, V.; Moss, S. E., Annexins: From structure to function. Physiol. Rev. 2002, 82, (2), 331-71.
- Saad, R. B.; Ben Romdhane, W.; Ben Hsouna, A.; Mihoubi, W.; Harbaoui, M.; Brini, F., Insights into plant annexins function in abiotic and biotic stress tolerance. Plant Signal Behav. 2020, 15, (1), 1699264.
- Laohavisit, A.; Davies, J. M., Annexins. New Phytol. 2011, 189, (1), 40-53.
- Wu, X.; Wang, Y.; Bian, Y.; Ren, Y.; Xu, X.; Zhou, F.; Ding, H., A critical review on plant annexin: Structure, function, and mechanism. Plant Physiol. Biochem. 2022, 190, 81-89.
- Yuan, F.; Yang, H.; Xue, Y.; Kong, D.; Ye, R.; Li, C.; Zhang, J.; Theprungsirikul, L.; Shrift, T.; Krichilsky, B.; Johnson, D. M.; Swift, G. B.; He, Y.; Siedow, J. N.; Pei, Z. M., Osca1 mediates osmotic-stress-evoked ca2+ increases vital for osmosensing in arabidopsis. Nature. 2014, 514, (7522), 367-71.
- Li, Y.; Yuan, F.; Wen, Z.; Li, Y.; Wang, F.; Zhu, T.; Zhuo, W.; Jin, X.; Wang, Y.; Zhao, H.; Pei, Z. M.; Han, S., Genome-wide survey and expression analysis of the osca gene family in rice. BMC Plant Biol. 2015, 15, 261.
- Ding, S.; Feng, X.; Du, H.; Wang, H., Genome-wide analysis of maize osca family members and their involvement in drought stress. PeerJ. 2019, 7, e6765.
- Yang, X.; Xu, Y.; Yang, F.; Magwanga, R. O.; Cai, X.; Wang, X.; Wang, Y.; Hou, Y.; Wang, K.; Liu, F.; Zhou, Z., Genome-wide identification of osca gene family and their potential function in the regulation of dehydration and salt stress in gossypium hirsutum. J. Cotton Res. 2019, 2, (1), 11.
- Haswell, E. S.; Meyerowitz, E. M., Mscs-like proteins control plastid size and shape in arabidopsis thaliana. Curr. Biol. 2006, 16, (1), 1-11.
- Hamilton, E. S.; Haswell, E. S., The tension-sensitive ion transport activity of msl8 is critical for its function in pollen hydration and germination. Plant Cell Physiol. 2017, 58, (7), 1222-1237.
- Peyronnet, R.; Haswell, E. S.; Barbier-Brygoo, H.; Frachisse, J. M., Atmsl9 and atmsl10: Sensors of plasma membrane tension in arabidopsis roots. Plant Signal. Behav. 2008, 3, (9), 726-9.
- Li, J.; Zhang, M.; Sun, J.; Mao, X.; Wang, J.; Wang, J.; Liu, H.; Zheng, H.; Zhen, Z.; Zhao, H.; Zou, D., Genome-wide characterization and identification of trihelix transcription factor and expression profiling in response to abiotic stresses in rice (oryza sativa l.). Int. J. Mol. Sci. 2019, 20, (2), 251.
- Finka, A.; Cuendet, A. F.; Maathuis, F. J.; Saidi, Y.; Goloubinoff, P., Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell. 2012, 24, (8), 3333-48.
- Knight, H., Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 2000, 195, 269-324.
- Mohanta, T. K.; Yadav, D.; Khan, A. L.; Hashem, A.; Abd Allah, E. F.; Al-Harrasi, A., Molecular players of ef-hand containing calcium signaling event in plants. Int. J. Mol. Sci. 2019, 20, (6), 1476.
- Braam, J.; Davis, R. W., Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in arabidopsis. Cell. 1990, 60, (3), 357-364.
- Al-Quraan, N. A.; Locy, R. D.; Singh, N. K., Expression of calmodulin genes in wild type and calmodulin mutants of arabidopsis thaliana under heat stress. Plant Physiol. Biochem. 2010, 48, (8), 697-702.
- Choi, M. S.; Kim, M. C.; Yoo, J. H.; Moon, B. C.; Koo, S. C.; Park, B. O.; Lee, J. H.; Koo, Y. D.; Han, H. J.; Lee, S. Y.; Chung, W. S.; Lim, C. O.; Cho, M. J., Isolation of a calmodulin-binding transcription factor from rice (oryza sativa l.). J. Biol. Chem. 2005, 280, (49), 40820-31.
- Hardoim, P. R.; de Carvalho, T. L.; Ballesteros, H. G.; Bellieny-Rabelo, D.; Rojas, C. A.; Venancio, T. M.; Ferreira, P. C.; Hemerly, A. S., Genome-wide transcriptome profiling provides insights into the responses of maize (zea mays l.) to diazotrophic bacteria. Plant Soil. 2020, 451, 121-143.
- 吉福德,J.L.;沃尔什,MP;Vogel, H. J., 结构和金属离子结合特性2+-绑定螺旋-环-螺旋-手绘图案。生化. J. 2007, 405, (2), 199-221.
- 昆杜,P.;尼赫拉,A.;吉尔,R.;图特哈,N.;Gill, S. S., 揭示植物中 ef-hand 介导的钙信号传导的重要性。S. Afr. J. Bot. 2022, 148, 615-633.
- 牛伟棠;韩,X.-W.;魏,S.-S.;尚志玲;王杰;杨,D.-W.;范,X.;高,F.;郑 S.-Z.;白,J.-T.;张,B.;王志伟;李,B.;Napier,R.,拟南芥环核苷酸门控通道6在热休克期间被多种钙调蛋白亚型负调控。J. Exp. 机器人。 2020, 71, (1): 90-104.
- 麦科马克,E.;蔡永昌;Braam,J.,处理钙信号传导:拟南芥凸轮和cmls。趋势 植物科学 2005, 10, (8): 383-9.
- 阿蒂夫,RM;沙希德,L.;瓦卡斯,M.;阿里,B.;拉希德,M.A.R.;阿泽姆,F.;马萨诸塞州纳瓦兹;瓦尼,SH;Chung, G., 关于钙依赖性蛋白激酶 (cpks) 信号传导对植物非生物胁迫耐受性的见解。国际 J. Mol. Sci. 2019, 20, (21): 5298.
- 叶德洛梅尔,T.;Boudsocq,M.,拟南芥中钙依赖性蛋白激酶及其亲缘物的性质和功能。新植物醇。 2019, 224, (2): 585-604.
- 胡, Z.;吕,X.;夏旭;周, J.;石,K.;俞,J.;周,Y.,番茄钙依赖性蛋白激酶的全基因组鉴定和表达分析。前面。植物科学 2016, 7, 469.
- 张国强;韩永棠;赵F.L.;胡,Y.;高永瑞;马,Y.F.;郑永;王永杰;温, Y. Q., 葡萄CDPK基因家族的全基因组鉴定和表达分析BMC植物生物学 2015, 15, 164.
- 张H.;刘伟忠;张永;邓,M.;牛F.;杨,B.;王旭;王,B.;梁,W.;德霍洛斯,MK;江,Y.Q.,油菜(芸苔型油菜)钙依赖性蛋白激酶(cpk)基因的鉴定,表达和相互作用分析。BMC 基因。 2014, 15, 211.
- 吴蒲;王伟;段,W.;李英;侯旭, 大白菜cdpk-snrk超家族基因综合分析及其在植物中的进化意义.前面。植物科学 2017, 8, 162.
- 徐, X.;刘,M.;卢,L.;他,M.;瞿伟;徐庆;齐,X.;Chen, X., 黄瓜中钙依赖性蛋白激酶基因家族的全基因组分析和表达。摩尔·热内特。基因。2015, 290, (4), 1403-14.
- 蔡,H.;程,J.;闫,Y.;肖,Z.;李杰;牟氏;邱,A.;赖,Y.;关,D.;He, S., 辣椒钙依赖性蛋白激酶及其密切相关激酶基因的全基因组鉴定和表达分析.前面。植物科学 2015, 6, 737.
- 刘杰;Zhu, J. K., 植物耐盐性所需的钙传感器同源物。科学。 1998, 280, (5371), 1943-1945.
- Luan, S., 植物钙信号转导中的 cbl-cipk 网络。趋势 植物科学 2009, 14, (1): 37-42.
- Sanyal,SK;马希瓦尔,S.;南比亚尔,DM;Pandey, G. K., Cbl-cipk 模块介导的磷酸化调节:事实和假设。生化. J. 2020, 477, (5): 853-871.
- 他,H.;加西亚-马塔,C.;He, L.-F., 植物生理反应中硫化氢与激素的相互作用.植物生长调节。 2018, 87, (1), 175-186.
- 杨,F.;董F.-s.;胡,F.-h.;刘永伟;柴,J.-f.;赵H.;吕,M.-y.;周,S.,小麦钙调蛋白结合转录激活因子(camta)基因家族的全基因组鉴定和表达分析(triticum aestivum l.)。BMC基因。 2020, 21, (1): 105.
- 基多科罗,S.;小浦,I.;索马,F.;筱崎,K.;铃木,T.;田仓,M.;宫川,T.;今泉,T.;Yamaguchi-Shinozaki, K., 时钟调节的共激活因子选择性地控制基因表达,以响应拟南芥中不同的温度胁迫条件。Proc. Natl. Acad. Sci. . 2023, 120, (16), e2216183120.
- 李志光;龚,M.;谢,H.;杨,L.;Li, J., 硫化氢供体硫氢化钠诱导的烟草 (nicotiana tabacum l.) 悬浮培养细胞的耐热性和 ca 的参与2+和钙调蛋白。植物科学 2012, 185, 185-189.
- 王玲玲;万,R.;石英;Xue, S., 硫化氢通过 ost1 和 ca 激活 s 型阴离子通道2+模块。摩尔植物。 2016, 9, (3): 489-491.
- 克雷布斯,J.;阿杰隆,LB;米哈拉克,M.,加利福尼亚州2+体内平衡和内质网 (ER) 应激:钙信号传导的综合视图。生化。生物物理学。Res. Commun.2015, 460, (1), 114-121.
- 卡雷拉斯-苏雷达,A.;皮汉,P.;Hetz, C.,内质网的钙信号传导:微调应激反应。细胞钙。 2018, 70, 24-31.
