Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Hui Li and Version 2 by Fanny Huang.
In cis-regulatory elements, enhancers and promoters with complex molecular interactions are used to coordinate gene transcription through physical proximity and chemical modifications. These processes subsequently influence the phenotypic characteristics of an organism. An in-depth exploration of enhancers and promoters can substantially enhance reseaourchers' understanding of gene regulatory networks, shedding new light on mammalian development, evolution and disease pathways.
- cis-regulatory elements
- enhancers
- promoters
1. Introduction
Unveiling the inherent laws of life is one of the main research objectives of modern biological science, and deciphering the regulatory mechanisms of phenotypes to investigate their associations with animal development, evolution and disease is of paramount importance in contemporary biological research directions. Cis-regulatory elements (CREs), comprising enhancers and promoters (EPs) and based on their chromatin three-dimensional spatial structure, regulate the precise initiation and efficiency of gene transcription by binding with transcription factors (TFs). These play an indispensable role in the regulation of gene expression. Within organisms, development is a highly ordered and dynamic process, involving the temporal and spatial regulation of numerous genes. EPs can interact with regulatory factors, orchestrating gene expression across various developmental stages. EPs have also played pivotal roles during mammalian evolution. By comparing EP sequences from different mammals, conserved and variable regions in these sequences have been identified, further shedding light on the evolutionary dynamics of mammalian gene regulatory networks and uncovering the genetic differences between mammalian species. With respect to diseases, EPs have been associated with cancers, neurological disorders and autoimmune diseases. Exposing their roles in disease mechanisms promises to offer new molecular targets for the development of innovative therapeutic methods.
2. Cis-Regulation Element
Enhancers are short DNA sequences within the genome that, by binding with various TF and cofactors, act upon promoters to regulate gene transcription. Enhancers operate without being limited by either direction or distance and are not restricted to the regulation of any specific gene. A single enhancer, influenced by the three-dimensional conformation of chromatin, often regulates the transcription of multiple genes [1].
In mammalian cells, active enhancer DNA sequences are loose and open, and they provide the environmental conditions for enhancers to transcribe genes [2]. Studies have shown that RNA polymerase II (Pol II) acts on promoters while also inducing the transcription of enhancers, resulting in the production of enhancer RNA (eRNA) [3]. ERNAs usually have short half-lives, low abundance and they lack RNA processing ability. They were initially regarded as sequencing noise and a by-product of transcription [4]. In mammals, the C-terminal domain of the largest subunit of Pol II contains Ser5, which can be in the phosphorylated state of Ser5 and subsequently initiate transcription [5]. In addition, phospho-Pol II (Ser5P) has been shown to be enriched in enhancers [6]. The insertion of transcription terminators into the globin genes enhancer (HS2 enhancer) stops the formation of eRNA [7]. Interference with eRNAs with small interfering and short hairpin RNAs reduces the transcription of its enhancer target genes [8]. This evidence not only robustly confirms the existence of eRNAs but also signifies their crucial role in the regulation of enhancer target genes. Hence, active enhancers can transcribe and generate eRNAs (Figure 1) and may regulate the transcription of target genes through eRNAs.
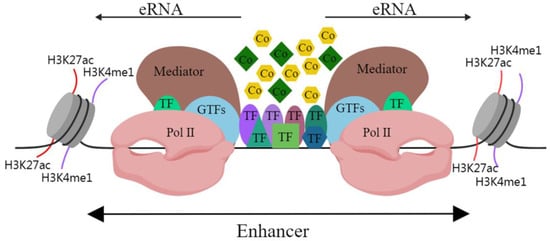
Figure 1. Schematic diagram of enhancer structure. Abbreviations: H3K27ac, acetylation of lysine 27 on histone 3; H3K4me1, monomethylation on lysine 4 of histone H3; TF, transcription factor; GTF, general transcription factor; Pol II, RNA polymerase II; Co, cofactor. The black line indicates DNA.
A promoter is a DNA sequence that allows a specific RNA transcript to be transcribed. The structure of the promoter affects its affinity for binding with RNA polymerase, thus affecting transcription levels. The structure of eukaryotic promoters typically consists of three parts: the core, proximal and distal regions [9]. Sequence-wise, the core promoter sequence contains the transcription start site (TSS). The vicinity of the TSS usually binds with general transcription factors (GTFs), Pol II and the mediator, leading to the formation of the pre-initiation complex (PIC). This region is termed the “core promoter” area, usually residing within 50 bp upstream and downstream of the TSS. It serves to determine the precise location and direction of transcription [10].
Core promoters are not structurally fixed, but classical elements include the TFIIB recognition element (BRE), TATA box, initiator (INR) and downstream promoter element (DPE). The BRE often accompanies the TATA box, and it is located either upstream or downstream of this and can significantly influence the binding of RNA polymerase to it [11]. The TATA box usually appears approximately 25 bp upstream of the initiation of transcription and it determines the onset of this process. It is also a binding site for RNA polymerase [12]. The INR element, encompassing the transcription initiation site, is a common functional element in the promoter of many protein-coding genes [13], playing a vital role in the initiation of transcription [14]. Moreover, core promoters containing the DPE element often lack the TATA box element [15]. The function of the DPE is similar to that of the TATA box and when it is mutated, it directly affects gene transcription [16]. The complexity of promoter sequences mostly lies in the core promoter, potentially due to the diverse and interchangeable GTFs that bind with it. These enable the precise recognition of various core promoter sequences [17]. The surrounding sequences of the core promoter also bind with TFs, recruiting cofactors that impact on the pre-initiation complex bound to the core promoter, which subsequently influence the initiation and elongation of transcription [18]. The regions around the core promoter that can bind to TFs, and based on the proximity to the gene, are termed the “proximal promoter” and “distal promoter” areas, which are collectively referred to as the “promoter” (Figure 2) [19].
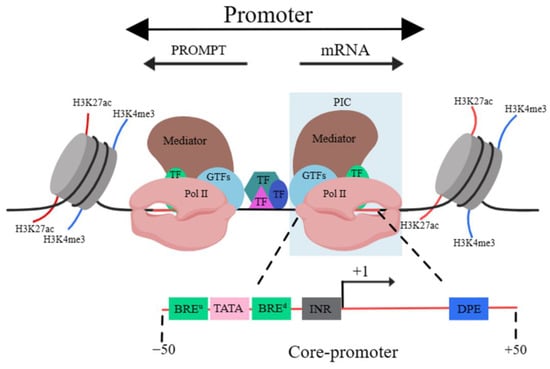
Figure 2. Schematic diagram of promoter structure. Abbreviations: H3K27ac, acetylation of lysine 27 on histone 3; H3K4me3, Trimethylation of histone H3 lysine 4. TF, transcription factor; GTF, general transcription factor; Pol II, RNA polymerase II; PIC, pre-initiation complex; BREu, BRE upstream (of TATA box); BREd, BRE downstream (of TATA box); Inr, initiator element; DPE, downstream promoter element. The black line indicates DNA.
It should be mentioned that the TSS of the majority of genes are in pairs. Thus, there is another TSS in the proximal upstream region of the opposite strand. This implies that promoters can undergo “divergent transcription”, producing promoter upstream transcripts (PROMPTs)/upstream antisense RNAs (uaRNAs) and mRNAs. PROMPTs are typically shorter than 500 bp, are not usually spliced and can be readily degraded by exosomes [20].
