Please note this is a comparison between Version 2 by Mona Zou and Version 1 by Sanjay Gupta.
The Melanoma Antigen Gene (MAGE) belongs to the larger family of cancer testis antigens. The MAGEA family were the first tumor-associated antigens identified at the molecular level whose expression was consistent in most human cancers and germinal cells. Aberrant expression of MAGEA family is noted in a majority of human malignancies, where they are associated with increased cancer cell proliferation, survival, and resistance to various therapies. This makes them an ideal biomarker and attractive therapeutic target in designing novel therapies.
- bladder cancer
- Melanoma Antigen Gene
- cancer testis antigen
- biomarkers
- therapeutic target
- signaling pathways
- protein–protein interaction
- genomic alterations
1. Expression of MAGEA Family in Bladder Cancer
There are a few reports on the expression of specific MAGEA family members in bladder cancers. Patard et al. (1995) analyzed the expression of MAGEA1, MAGEA2, MAGEA3, and MAGEA4 and observed the expression of at least one of these genes in 61% of invasive and 28% of superficial tumors, with MAGEA3 and MAGEA4 genes being most frequently expressed [53][1]. A study by Picard et al. (2007) analyzed the expression of MAGEA3, MAGEA4, MAGEA8, and MAGEA9 in bladder cancer. The study reported that MAGEA3–9 members were expressed in 30%, 33%, 56%, and 54% of bladder tumors, respectively. Although MAGEA8 was the most frequently expressed, its expression was low overall and mostly confined to the normal urothelium. In comparison, MAGEA9 was expressed at a higher level and was two times more frequent in superficial bladder cancer than in invasive tumors [54][2]. Bergeron et al. (2009) showed that MAGEA4 and MAGEA9 were expressed in 38% and 63% of NMIBC, in 48% and 57% of MIBC, 65% and 84% in carcinomas in situ, and 73% and 85% in lymph node metastases, respectively. The expression of MAGEA4 (p = 0.007) and MAGEA9 (p = 0.012) was associated with higher-grade tumors. In multivariable Cox regression analyses, the expression of MAGEA9 in pTa tumors was associated with recurrence (HR = 1.829; p = 0.010). MAGEA4 expression in these tumors was associated with progression to MIBC (HR = 7.417, p = 0.013) based on univariate analyses, whereas MAGEA9 expression was further predictive of bladder cancer progression [55][3]. A study by Xylinas et al. (2014) showed that MAGEA3 expression was independently associated with an increased risk of bladder cancer recurrence and cancer-specific mortality [56][4]. Another study by Dyrskjot et al. (2012) showed that 43% of bladder tumors expressed MAGEA3. A univariate Cox regression analysis of gene expression in NMIBC showed that the expression of MAGEA3 (p = 0.026) was significantly associated with a shorter progression-free survival [57][5].
The researchers performed a meta-analysis combining the results from several independent studies on bladder cancer listed in the TCGA database for detecting differentially expressed MAGEA genes with the potential to increase both the statistical power and generalizability of their analysis. MAGEA family expression was analyzed in n = 4536 patients according to tumor stage and the p values were generated by two-sample t-test, comparing pTa and pT2 stage bladder cancer. MAGEA2 (p < 0.001), MAGEA3 (p = 0.005), MAGEA6 (p < 0.001), and MAGEA12 (p = 0.01) were found to have stage-dependent expression (highest in pT2 subgroup), suggesting that these MAGEA members might play important roles in the development of urothelial carcinoma (Figure 21). These observations highlight the importance of improving their understanding of the etiology of bladder cancer, as well as the molecular changes underlying aberrant MAGEA expression. However, the clinical and prognostic value of MAGEA family members in the pathobiology of bladder cancer is currently under investigation by their group.
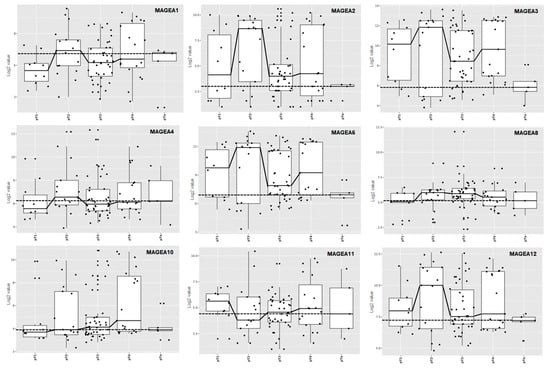
Figure 21. MAGEA expression profile across various stages of bladder cancer. Box and dot-plot of bladder cancer stages. The X-axis of the graph represents various stages of bladder cancer, pT1, pT2, pT3, pT4, and pTa, and the Y-axis of the graph represents log2 value.
2. Genomic Aberration of MAGEA Genes in Bladder Cancer
To gain a better understanding of the molecular alteration of MAGEA proteins in bladder cancer, the researchers used a TCGA bladder cancer patient’s (n = 4536) genomics database to analyze how MAGEA family members could affect bladder tumorigenesis. Genomic alterations, including mutation, homozygous deletion, or amplification, led to uncontrolled proliferation and irregularities in cell death in neoplastic cells [64,65][6][7]. It was found that frequencies of genomic alterations among MAGEA family members in bladder cancer were MAGEA1 1.8%, MAGEA2 1.6%, MAGEA3 1.9%, MAGEA4 1.6%, MAGEA6 2.6%, MAGEA8 1.7%, MAGEA10 2%, MAGEA11, 2.1%, and MAGEA12 2.4% (Figure 32A). The most prevalent form of genomic alteration is gene amplification among all MAGEA members [66,67][8][9]. The amplification of these genes often has the potential to transform normal cells into neoplastic cells, further hinting at the possibility that the MAGEA family has a significant role in bladder tumorigenesis [68][10]. These observations recommend that an analysis of MAGEAs might be a useful diagnostic tool to determine the invasive potential of bladder cancer. In addition, the amplified regions of some MAGEA genes might serve as potential therapeutic targets.
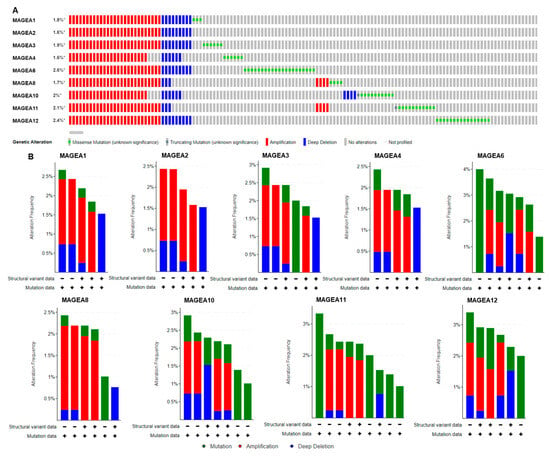
Figure 32. Genomic alterations in MAGEA family members. Oncoprint of MAGEA1, MAGEA2, MAGEA3, MAGEA4, MAGEA6, MAGEA8, MAGEA10, MAGEA11, and MAGEA12 in bladder cancer patients (n = 5436). (A) Color bar represents the individual patient’s profile showing gene amplification in red, * denotes % gene amplification; deletion in blue; and mutation in green. (B) Bar graph represents the summary of MAGEA gene alteration frequency (Y-axis), and the X-axis shows the mutation rate (green), amplification (red), and deep deletion (blue) in bladder cancer patients.
The researchers also analyzed mutations in the MAGEA family in bladder cancer. MAGEA genes are more frequently mutated in cancer patients, suggesting their critical role in malignancy [69][11]. In bladder cancer, the somatic mutation frequency is very low and ranges from 0.1% to 0.4% in the MAGEA family members identified in 4880 samples from 4158 patients consolidated from 21 studies from the cancer genome atlas (TCGA) (Figure 32B).
Moreover, missense mutations were identified in MAGEA6 compared to MAGEA3 and MAGEA11. Some of these mutations, including G137W, E232Q, P242L, Y249H, P262R, G296V, R298C, and E314Q, were found in the 229–399 amino acid sequence in the MAGEA6 gene [56][4]. Among the noted mutations, the functional impact of P262R was most deleterious, which might contribute to poor outcomes in bladder cancer patients (Figure 43A,B).
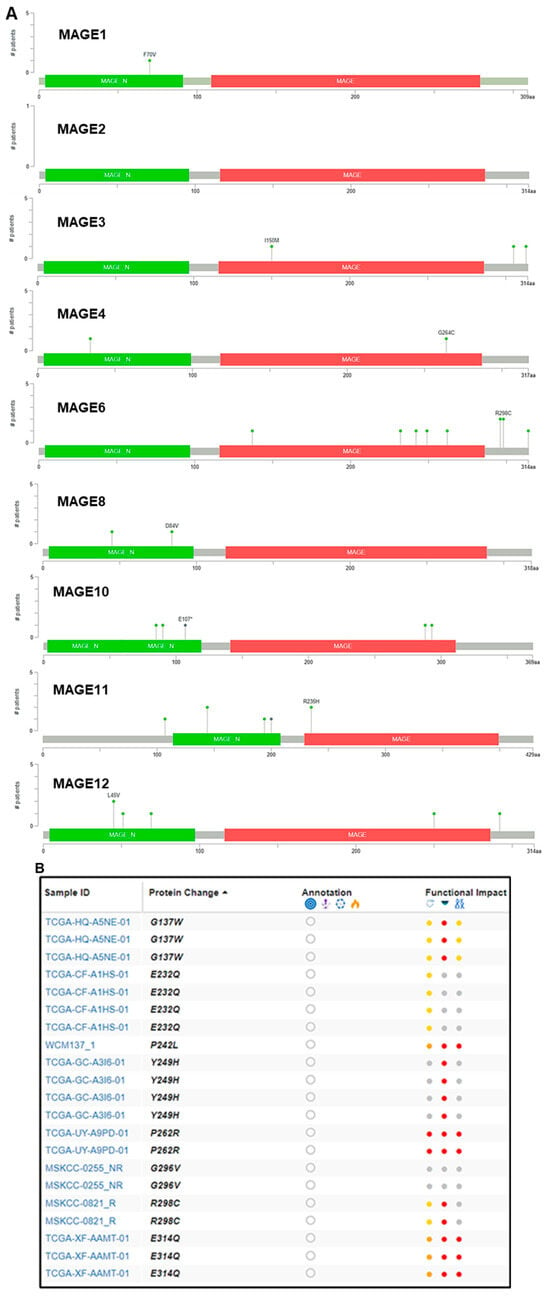
Figure 43. Mutation in the MAGEA family in bladder cancer. (A) Mutation plot was generated by the Mutation Mapper tool (cBioportal) showing the structure of MAGEA protein, and the frequency and position of mutations. Green color shows MAGE_N: Melanoma-associated antigen family N terminal (4–97), and red color shows MAGE: MAGE family (116–286). The green lollipop denotes number and change in the amino acid. (B) The table shows the patient’s TCGA sample ID, protein change, functional impact, and mutation type. Color dots denotes function impact: red—high, yellow/orange—moderate, and gray—low impact.
3. Gene Network and Signaling Pathways of MAGEA Family in Bladder Cancer
The altered expression of genes results in changes in gene expression and gene network interaction during cancer progression. A Cytoscape version 3.10.1 (Complex Network Analysis) Genemania module [70][12] was used to explore the genetic interaction of MAGEA1, MAGEA2, MAGEA3, MAGEA4, MAGEA6, MAGEA8, MAGEA10, MAGEA11, and MAGEA12 (red color). This software tool provides a critical assessment and integration of protein–protein interactions to assess the associations of potential differentially expressed genes. The size of the circle of each protein represents its degree of connection to other proteins. The analysis demonstrated the interaction of MAGEA family members with other MAGE members, including MAGEA2B, MAGEB10, MAGEB2, and others (blue color). The available scientific literature reports on the interaction between MAGEA and MAGEB in bladder cancer [71][13]. Moreover, both family members share the MAGE domain that influences the tumor microenvironment and promotes cell proliferation (Figure 54A).
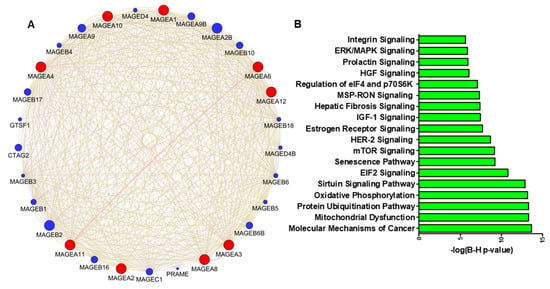
Figure 54. (A) Protein–protein interaction network of MAGEA family members constructed by Cytoscape software (version 3.10.1). Proteins are represented with color nodes, and interactions are represented with edges. The size of the circle of each protein represents its connection degree to other proteins. The red color circle shows the query protein, and the blue color shows the interacting proteins. (B) Signaling pathways associated with MAGEA family; the X-axis shows the log(B-H) value and the Y-axis represents the associated pathway in bladder cancer.
4. MAGEA Family as Diagnostic Biomarkers in Bladder Cancer
Several studies have analyzed the stage-specific expression of the MAGEA family in bladder cancer, indicating their higher levels in invasive disease [53,54,55,56,57,58,59,60,61,62,63][1][2][3][4][5][14][15][16][17][18][19]. Therefore, the researchers explored the TCGA database to determine the expression of different MAGEA members in bladder cancer (BCLA; n = 414) compared with normal bladder samples (n = 19). The BCLA patient’s dataset was not available for MAGEA9, and MAGEA5 and MAGEA7 were excluded from the analysis as they are pseudogenes. In some databases, the expression of MAGEA5 and MAGEA7 are measured, despite being pseudogenes, as they regulate oncogenes by serving as miRNA decoys [73][20]. Interestingly, MAGEA2, MAGEA3, MAGEA4, MAGEA6, MAGEA10, MAGEA11, and MAGEA12 exhibited significant differences in their expression in bladder cancer compared to normal bladder samples. Other MAGEA family members, including MAGEA1 and MAGEA8, exhibited lower expression in bladder cancer compared to normal bladder specimens (Figure 65). A data analysis of bladder cancer patients further uncovers the possibilities of utilizing MAGEA2, MAGEA3, MAGEA4, MAGEA6, MAGEA10, MAGEA11, and MAGEA12 members as diagnostic biomarkers for bladder cancer.
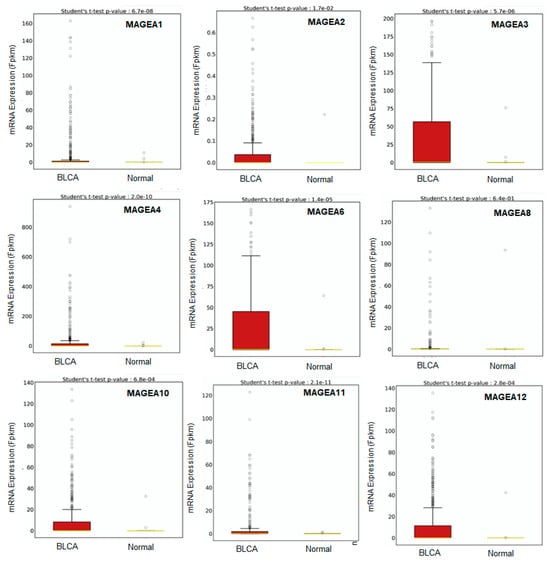
Figure 65. Transcriptomic expression of MAGEA family. Relative expressions of varying MAGEA types in normal samples and bladder cancer samples (BLCA) quantified through a box plot analysis of the TCGA dataset. The X-axis shows the expression of 435 patients (BCLA = 414, and Normal = 19), and the Y-axis shows the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) mRNA expression values.
Next, the researchers determined the cancer-specific mortality using a hazard ratio (HR) of MAGEA proteins with overall survival for the assessment of relative risk for bladder cancer aggressiveness, using the GENT2 database, which associates gene expression with the HR. The HR was calculated using the fixed effect model and the random effect model with 95% CI and p-value. Based on the data analysis from various studies, the HR value for the fixed (FE) and random effect (RE) models were as follows: MAGEA1 (FE, 1.06; RE 1.18; p-value = 0.07); MAGEA2 (FE, 1.48; RE, 1.48; p-value = 0.67); MAGEA3 (FE,1.43; RE, 1.43; p-value = 0.33); MAGEA4 (FE, 0.96; RE, 1.21; p-value = 0.02); MAGEA6 (FE, 0.99, RE, 1.05; p-value = 0.03); MAGEA8 (FE, 1.05, RE, 1.05; p-value = 0.86); MAGEA10 (FE, 1.07.; RE,1.10; p-value = 0.11); MAGEA11 (FE, 1.09; RE,0.94; p-value = 0.31); and MAGEA12 (FE, 1.00; RE, 1.73; p-value < 0.01). The HR ratio of MAGEA6 and MAGEA12 was statistically significant with p-value = 0.03; <0.01. Based on these findings, poor survival might be predicted in patients that express elevated levels of MAGEA6 and MAGEA12. To define the variation among different datasets, the heterogeneity was calculated. Heterogeneity was common regardless of the treatment effects by odds ratios or risk differences. Random effects estimates, which incorporate heterogeneity, tended to be less precisely assessed than fixed effects estimates. Therefore, compared with the fixed effect model, the weights assigned under random effects are more balanced. The heterogeneity was 0%; this means that heterogeneity has no importance in the results displayed in the forest plot (Figure 76).
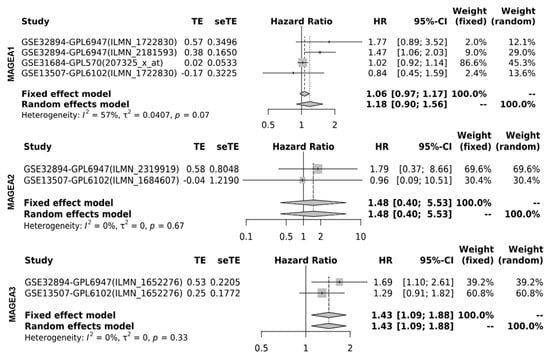
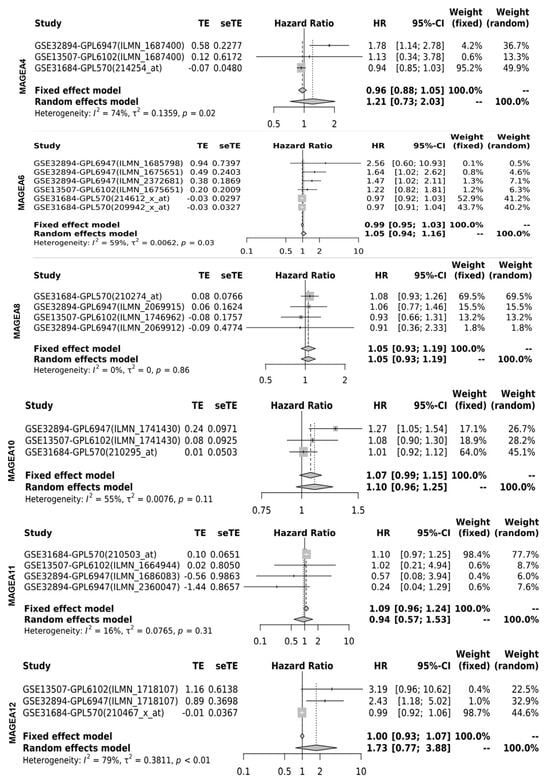
Figure 76. Hazard ratio (HR) of MAGEA proteins in bladder cancer. Forest plots were generated for hazard ratio in bladder cancer patients. HR was calculated using the fixed effect model and the random effect model with 95% CI with the p-value of heterogeneity.
5. Prognostic Value of MAGEA Gene Family in Bladder Cancer
The researchers analyzed the prognostic value (log rank) of MAGEA family members viz. MAGEA -1, -3, -4, -6, -8, -10, -11, and -12 in bladder cancer patients (n = 408; MAGEA2, n = 406) using KM-Plot (http://kmplot.com/analysis (accessed on 19 October 2023)). For this, the researchers performed survival analysis in bladder cancer patients using the TCGA database. A hazard ratio (HR) of more than 1.0 was used to predict the potential of genes as prognostic biomarkers. Based on the log rank value of MAGEA family members, the researchers found that MAGEA6 scored the highest log rank (log rank 0.99), followed by MAGEA3 (log rank 0.83), compared to other MAGEA members. These values suggest that MAGEA3 and MAGEA6 may serve as prognostic biomarkers for bladder cancer patients (Figure 87). Additional clinical validation in bladder cancer patients is required to justify the above rationale.
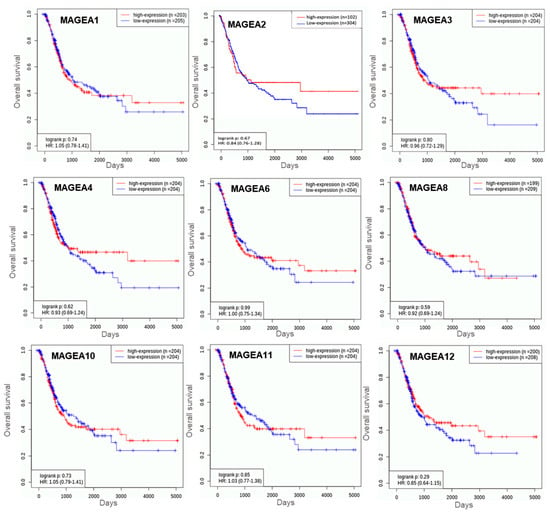
Figure 87. Kaplan–Meier plots for overall survival in bladder cancer. These curves were generated using the TCGA bladder cancer database of n = 408 patients. The X-axis shows the overall survival in days and the Y-axis shows the overall survival rate.
Next, the researchers determined the predicted location and protein–protein interaction of MAGEA3 and MAGEA6 genes in bladder cancer. MAGEA6 protein is intracellular in location and showed direct interaction (physical association) with secreted (intracellular) protein S100A9, CTF1, GFOD1, and APO4. MAGEA6 showed interaction with intracellular proteins such as TULP3, EXOC5, LSM2, and others. Similarly, MAGEA3 showed interaction with secreted intracellular protein S100A9, along with other intracellular proteins. Genomic association revealed a direct correlation of MAGEA3 and MAGEA6 with calcium-binding protein S100A9 (Figure 98A,B). To further understand the association between MAGEA3 and MAGEA6 with S100A9, the researchers explored the OncoDB cancer database, a large-scale multi-omics database, and performed pairwise gene expression correlation analysis between MAGEA3 and MAGEA6 and S100A9 in bladder cancer patients (Figure 109). S100A9 is a secretory protein, and its expression is positively associated with elevated levels of MAGEA3 and MAGEA6 proteins [74,75][21][22]. An overexpression of S100A9 is associated with stage progression, invasion, metastasis, and poor survival in bladder cancer patients [76][23]. S100A9 and MAGEA family members may be cooperative oncogenes, given their findings.
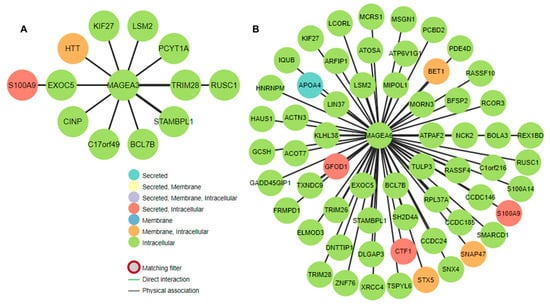
Figure 98. Gene interaction of (A) MAGEA3 and (B) MAGEA6 in bladder cancer. The figure shows the physical association and gene regulatory relationships. The assorted colors of the nodes show the predicted location of the proteins.
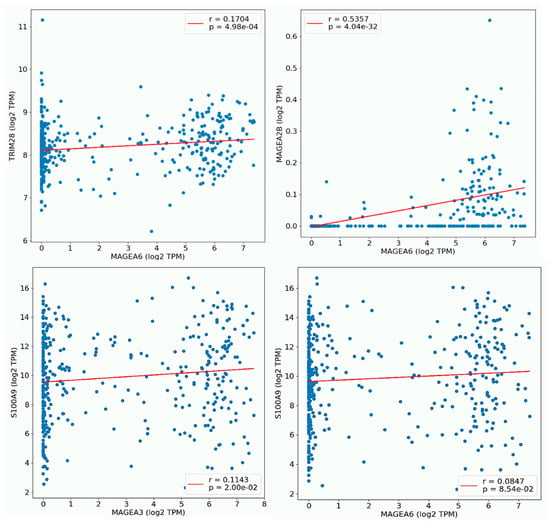
Figure 109. Plot pair-wise gene expression correlation analysis between MAGEA6, with TRM28, and MAGEA28. MAGEA3 showed its correlation with S100A9 in BLCA. The X and Y-axes of the graph show log2 transcript per million (TPM).
References
- Patard, J.J.; Brasseur, F.; Gil-Diez, S.; Radvanyi, F.; Marchand, M.; François, P.; Abi-Aad, A.; Van Cangh, P.; Abbou, C.C.; Chopin, D.; et al. Expression of MAGE genes in transitional-cell carcinomas of the urinary bladder. Int. J. Cancer 1995, 64, 60–64.
- Picard, V.; Bergeron, A.; Larue, H.; Fradet, Y. MAGE-A9 mRNA and protein expression in bladder cancer. Int. J. Cancer 2007, 120, 2170–2177.
- Bergeron, A.; Picard, V.; LaRue, H.; Harel, F.; Hovington, H.; Lacombe, L.; Fradet, Y. High frequency of MAGE-A4 and MAGE-A9 expression in high-risk bladder cancer. Int. J. Cancer 2009, 125, 1365–1371.
- Xylinas, E.; Cha, E.K.; Khani, F.; Kluth, L.A.; Rieken, M.; Volkmer, B.G.; Hautmann, R.; Küfer, R.; Chen, Y.T.; Zerbib, M.; et al. Association of oncofetal protein expression with clinical outcomes in patients with urothelial carcinoma of the bladder. J. Urol. 2014, 191, 830–841.
- Dyrskjøt, L.; Zieger, K.; Kissow Lildal, T.; Reinert, T.; Gruselle, O.; Coche, T.; Borre, M.; Ørntoft, T.F. Expression of MAGE-A3, NY-ESO-1, LAGE-1 and PRAME in urothelial carcinoma. Br. J. Cancer 2012, 107, 116–122.
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558.
- Chen, J.; Herlong, F.H.; Stroehlein, J.R.; Mishra, L. Mutations of Chromatin Structure Regulating Genes in Human Malignancies. Curr. Protein Pept. Sci. 2016, 17, 411–437.
- Kang, J.U.; Koo, S.H.; Jeong, T.E.; Kwon, K.C.; Park, J.W.; Jeon, C.H. Multitarget fluorescence in situ hybridization and melanoma antigen genes analysis in primary bladder carcinoma. Cancer Genet. Cytogenet. 2006, 164, 32–38.
- Karihtala, P.; Kilpivaara, O.; Porvari, K. Mutational signatures and their association with survival and gene expression in urological carcinomas. Neoplasia 2023, 44, 100933.
- Jungbluth, A.A.; Busam, K.J.; Kolb, D.; Iversen, K.; Coplan, K.; Chen, Y.T.; Spagnoli, G.C.; Old, L.J. Expression of MAGE-antigens in normal tissues and cancer. Int. J. Cancer 2000, 85, 460–465.
- Nasrah, S.; Radi, A.; Daberkow, J.K.; Hummler, H.; Weber, S.; Seaayfan, E.; Kömhoff, M. MAGED2 Depletion Promotes Stress-Induced Autophagy by Impairing the cAMP/PKA Pathway. Int. J. Mol. Sci. 2023, 24, 13433.
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504.
- Ottaviani, S.; Colau, D.; van der Bruggen, P.; van der Bruggen, P. A new MAGE-4 antigenic peptide recognized by cytolytic T lymphocytes on HLA-A24 carcinoma cells. Cancer Immunol. Immunother. CII 2006, 55, 867–872.
- Kocher, T.; Zheng, M.; Bolli, M.; Simon, R.; Forster, T.; Schultz-Thater, E.; Remmel, E.; Noppen, C.; Schmid, U.; Ackermann, D.; et al. Prognostic relevance of MAGE-A4 tumor antigen expression in transitional cell carcinoma of the urinary bladder: A tissue microarray study. Int. J. Cancer 2002, 100, 702–705.
- Mengus, C.; Schultz-Thater, E.; Coulot, J.; Kastelan, Z.; Goluza, E.; Coric, M.; Spagnoli, G.C.; Hudolin, T. MAGE-A10 cancer/testis antigen is highly expressed in high-grade non-muscle-invasive bladder carcinomas. Int. J. Cancer 2013, 132, 2459–2463.
- Mohsenzadegan, M.; Razmi, M.; Vafaei, S.; Abolhasani, M.; Madjd, Z.; Saeednejad Zanjani, L.; Sharifi, L. Co-expression of cancer-testis antigens of MAGE-A6 and MAGE-A11 is associated with tumor aggressiveness in patients with bladder cancer. Sci. Rep. 2022, 12, 599.
- Lerut, E.; Van Poppel, H.; Joniau, S.; Gruselle, O.; Coche, T.; Therasse, P. Rates of MAGE-A3 and PRAME expressing tumors in FFPE tissue specimens from bladder cancer patients: Potential targets for antigen-specific cancer immunotherapeutics. Int. J. Clin. Exp. Pathol. 2015, 8, 9522–9532.
- Bar-Haim, E.; Paz, A.; Machlenkin, A.; Hazzan, D.; Tirosh, B.; Carmon, L.; Brenner, B.; Vadai, E.; Mor, O.; Stein, A.; et al. MAGE-A8 overexpression in transitional cell carcinoma of the bladder: Identification of two tumour-associated antigen peptides. Br. J. Cancer 2004, 91, 398–407.
- Sharma, P.; Shen, Y.; Wen, S.; Bajorin, D.F.; Reuter, V.E.; Old, L.J.; Jungbluth, A.A. Cancer-testis antigens: Expression and correlation with survival in human urothelial carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 5442–5447.
- Tang, Z.; Wei, G.; Zhang, L.; Xu, Z. Signature microRNAs and long noncoding RNAs in laryngeal cancer recurrence identified using a competing endogenous RNA network. Mol. Med. Rep. 2019, 19, 4806–4818.
- Yasar, O.; Akcay, T.; Obek, C.; Turegun, F.A. Significance of S100A8, S100A9 and calprotectin levels in bladder cancer. Scand. J. Clin. Lab. Investig. 2017, 77, 437–441.
- Verma, S.; Shankar, E.; Lin, S.; Singh, V.; Chan, E.R.; Cao, S.; Fu, P.; MacLennan, G.T.; Ponsky, L.E.; Gupta, S. Identification of Key Genes Associated with Progression and Prognosis of Bladder Cancer through Integrated Bioinformatics Analysis. Cancers 2021, 13, 5931.
- Kim, W.T.; Kim, J.; Yan, C.; Jeong, P.; Choi, S.Y.; Lee, O.J.; Chae, Y.B.; Yun, S.J.; Lee, S.C.; Kim, W.J. S100A9 and EGFR gene signatures predict disease progression in muscle invasive bladder cancer patients after chemotherapy. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 974–979.
More
 Encyclopedia
Encyclopedia
