Artificial intelligence (AI) has emerged as a cutting-edge tool, simultaneously accelerating, securing, and enhancing the diagnosis and treatment of patients. An exemplification of this capability is evident in the analysis of peripheral blood smears (PBS). In university medical centers, hematologists routinely examine hundreds of PBS slides daily to validate or correct outcomes produced by advanced hematology analyzers assessing samples from potentially problematic patients. This process may logically lead to erroneous PBC readings, posing risks to patient health. AI functions as a transformative tool, significantly improving the accuracy and precision of readings and diagnoses.
- deep learning (DP)
- blood cell classification
- “Naturalize” technique
- %MCEPASTEBIN%
1. Introduction
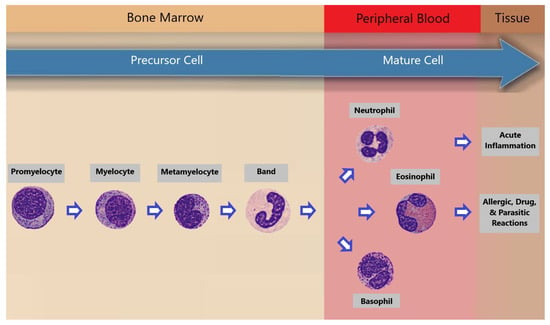
2. Peripheral Blood Smears Examination by Artificial Intelligence
Within the realm of machine learning, the conventional approach involved the manual extraction of image features followed by classification. The advent of deep learning revolutionized this process by automating image classification, particularly impactful in blood cell analysis. Previous studies primarily concentrated on five white blood cell (WBC) types: neutrophils, eosinophils, basophils, lymphocytes, and monocytes. Jung et al. [13][4] introduced W-Net, a CNN-based model for white blood cell (WBC) classification, achieving 97% accuracy with 10-fold cross-validation. Their work focused on classifying five WBC types: neutrophils, eosinophils, basophils, lymphocytes, and monocytes. Sahlol et al. [14][5] combined VGG-16 and a feature reduction algorithm (SESSA) to achieve 95.67% accuracy for WBC leukemia image classification. They categorized WBCs into five classes. Almezhghwi et al. [15][6] used ImageNet pre-trained architectures (VGG, ResNet, DenseNet) and data augmentation, with DenseNet-169 achieving 98.8% accuracy. Their study involved the classification of five WBC types. Tavakoli et al. [16][7] developed a segmentation algorithm and used SVM for WBC classification, achieving high accuracy in multiple datasets. They focused on five WBC types. Chen et al. [17][8] proposed a hybrid deep model combining ResNet and DenseNet with a spatial and channel attention module (SCAM), outperforming previous methods. They worked with datasets containing five WBC types. Katar et al. [18][9] applied transfer learning with ImageNet models, with MobileNet-V3-Small achieving 98.86% accuracy. They classified five WBC types. Nahzat et al. [19][10] designed a CNN-based model using the Kaggle BCCD dataset, achieving competitive results. They also focused on five WBC types. Heni et al. [20][11] introduced the EK-Means method for WBC image segmentation, achieving a validation accuracy of 96.24% with VGG-19. They categorized five WBC types. Ziquan Zhu [21,22][12][13] presented DLBCNet models that used ResNet-50 for feature extraction and achieved impressive accuracy. They worked with datasets containing five WBC types. Other groups of researchers [23,24,25,26,27,28][14][15][16][17][18][19] worked on the classification of 8 blood classes in the the PBC dataset [4][20]. Table 1 summarizes the work of those groups.| Paper | Year | Used Model | Classes | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|---|---|
| [23][14] | 2019 | Fine-Tuned VGG-16 | 8 | 96.20 | 97.00 | 96.00 | 97.00 |
| [24][15] | 2020 | ShuffleNet | 8 | 97.94 | 97.94 | 97.94 | 97.94 |
| [25][16] | 2021 | BloodCaps | 8 | 99.30 | 99.17 | 99.16 | 99.88 |
| [26][17] | 2021 | Ensemble ResNet-50-2 and VGG-19 | 8 | 99.51 | 99.47 | 99.58 | 99.93 |
| [27][18] | 2023 | CNN | 8 | 99.91 | 99.77 | 99.60 | 99.60 |
| [28][19] | 2023 | Mask R-CNN | 8 | 99.31 | 97.25 | 97.37 | 97.31 |
The “Naturalize” technique directly tackles two prominent challenges in deep learning: data insufficiency and class imbalance. In the realm of deep learning, the quality and balance of data significantly impact model performance. “Naturalize” effectively addresses these challenges by generating high-quality blood cell samples. This innovative approach substantially enhances the performance of blood cell classification, pushing the boundaries of deep learning in blood cell analysis. Not only does it improve accuracy, but it also ensures robustness and reliability when handling diverse and imbalanced blood cell classes.
References
- McKenzie, S.B.; Williams, L. Clinical Laboratory Hematology, 3rd ed.; Pearson: Upper Saddle River, NJ, USA, 2014.
- Rodak, B.F.; Carr, J.H. Clinical Hematology Atlas, 4th ed.; Saunders: Maryland Heights, MO, USA, 2012.
- Al-qudah, R.; Suen, C.Y. Peripheral blood smear analysis using deep learning: Current challenges and future directions. In Computational Intelligence and Image Processing in Medical Applications; World Scientific: Singapore, 2022; pp. 105–118.
- Jung, C.; Abuhamad, M.; Alikhanov, J.; Mohaisen, A.; Han, K.; Nyang, D. W-Net: A CNN-based architecture for white blood cells image classification. arXiv 2019, arXiv:1910.01091.
- Sahlol, A.T.; Kollmannsberger, P.; Ewees, A.A. Efficient classification of white Blood Cell Leukemia with improved swarm optimization of deep features. Sci. Rep. 2020, 10, 2536.
- Almezhghwi, K.; Serte, S. Improved classification of white blood cells with the generative adversarial network and deep convolutional neural network. Comput. Intell. Neurosci. 2020, 2020, 6490479.
- Tavakoli, S.; Ghaffari, A.; Kouzehkanan, Z.M.; Hosseini, R. New segmentation and feature extraction algorithm for classification of white blood cells in peripheral smear images. Sci. Rep. 2021, 11, 19428.
- Chen, H.; Liu, J.; Hua, C.; Feng, J.; Pang, B.; Cao, D.; Li, C. Accurate classification of white blood cells by coupling pre-trained ResNet and DenseNet with SCAM mechanism. BMC Bioinform. 2022, 23, 282.
- Katar, O.; Kilinçer, İ.F. Automatic classification of white blood cells using pre-trained deep models. Sak. Univ. J. Comput. Inf. Sci. 2022, 5, 462–476.
- Nahzat, S. White Blood Cell Classification Using Convolutional Neural Network. Technol. Eng. Res. 2022, 3, 32–41.
- Heni, A. Blood Cells Classification Using Deep Learning with customized data augmentation and EK-means segmentation. J. Theor. Appl. Inf. Technol. 2023, 101, 1162–1173. Available online: https://www.jatit.org/volumes/Vol101No3/11Vol101No3.pdf (accessed on 10 November 2020).
- Zhu, Z.; Wang, S.-H.; Zhang, Y.-D. ReRNet: A deep learning network for classifying blood cells. Technol. Cancer Res. Treat. 2023, 22, 15330338231165856.
- Zhu, Z.; Ren, Z.; Lu, S.; Wang, S.; Zhang, Y. DLBCNet: A deep learning network for classifying blood cells. Big Data Cogn. Comput. 2023, 7, 75.
- Acevedo, A.; Alférez, S.; Merino, A.; Puigví, L.; Rodellar, J. Recognition of peripheral blood cell images using convolutional neural networks. Comput. Methods Progr. Biomed. 2019, 180, 1050209.
- Ucar, F. Deep learning approach to cell classificatio in human peripheral blood. In Proceedings of the 2020 5th International Conference on Computer Science and Engineering (UBMK), Diyarbakir, Turkey, 9–11 September 2020.
- Long, F.; Peng, J.-J.; Song, W.; Xia, X.; Sang, J. BloodCaps: A capsule network based model for the multiclassification of human peripheral blood cells. Comput. Methods Progr. Biomed. 2021, 202, 105972.
- Gavas, E.; Olpadkar, K. Deep CNNs for peripheral blood cell classification. arXiv 2021, arXiv:2110.095082.
- Asghar, R.; Kumar, S.; Hynds, P.; Mahfooz, A. Automatic classification of blood cell images using convolutional neural network. arXiv 2023, arXiv:2308.06300.
- Atici, H.; Kocer, H.E. Mask R-CNN based segmentation and classification of blood smear images. Gazi Muhendis. Bilim. Derg. 2023, 9, 128–1433.
- Acevedo, A.; Merino, A.; Alférez, S.; Molina, Á.; Boldú, L.; Rodellar, J. A dataset of microscopic peripheral blood cell images for development of automatic recognition systems. Data Brief 2020, 30, 105474.
