Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Kívia Queiroz de Andrade and Version 2 by Sirius Huang.
The CCCH-type zinc finger antiviral protein (ZAP) in humans, specifically isoforms ZAP-L and ZAP-S, is a crucial component of the cell’s intrinsic immune response. ZAP acts as a post-transcriptional RNA restriction factor, exhibiting its activity during infections caused by retroviruses and alphaviruses.
- ZAP protein
- PARP-13
- antiviral activity
- Zinc Finger Antiviral Protein
1. Introduction
Organisms have several ways of sensing and controlling viral infections. This recognition occurs mainly through the detection of viral ribonucleic acid (RNA) or deoxyribonucleic acid (DNA) [1]. This triggers intracellular signaling events that ultimately result in the production of antiviral molecules [2]. In order for the virus to replicate successfully, it is crucial to evade the immune response of the host cell. As a result, the cell develops various recognition mechanisms and restriction factors to control infection. One of these restriction mechanisms is the degradation of viral RNA (vRNA). There are several cell intrinsic antiviral proteins that bind to RNA, regulate translation, and target it for decay, thereby interfering with different stages of the virus replication cycle [3]. Some of these restriction factors are induced by type I interferons [4], which create an antiviral state in neighboring cells. The human CCCH-type zinc finger antiviral protein (ZAP) acts as a post-transcriptional RNA restriction factor in the host cell for viruses such as retroviruses [5], filoviruses [6], alphaviruses [7]. ZAP also inhibits RNA translation and targets the vRNA for degradation [8][9][10][8,9,10]. ZAP is considered an interferon (IFN)-stimulated gene (ISG) that can be induced by viral infection [11]. It is capable of restricting several negative-sense single-stranded RNA viruses [12] and positive-sense single-stranded RNA viruses [9]; however, it is unclear what influence ZAP has on double-stranded RNA (dsRNA) viruses and other higher-order structured RNAs. ZAP was discovered as a protein with antiviral activity in rat cells that showed resistance to Moloney murine leukemia virus (MoMuLV or MMLV) infection [13]. In the presence of the ZAP protein, some viruses, such as herpes simplex virus type 1 (HSV-1), yellow fever virus (YFV) [7], Zika virus (ZIKV), and dengue virus (DENV) [10], are able to grow normally. Interestingly, this ability is not dependent on belonging to the same family, as coxsackievirus B3 [14] but not poliovirus [7], both from the Picornaviridae family, was inhibited by ZAP.
There are four isoforms of ZAP: the short isoform ZAP (ZAP-S), the medium isoform ZAP (ZAP-M), the long isoform ZAP (ZAP-L), and the extra-long isoform ZAP (ZAP-XL), with ZAP-S and ZAP-L being the most prevalent isoforms [15][16][15,16]. ZAP-L contains a poly(ADP-ribose) polymerase (PARP) domain and a CaaX motif, which undergoes S-farnesylation. This S-farnesylation is necessary for its antiviral activity against some viruses, such as Sindbis virus (SINV) [17][18][17,18]. This S-farnesylation appears to be the reason why ZAP-L is found in plasma membranes or membranous compartments (such as endolysosomes and endoplasmic reticulum) inside cells, while ZAP-S is found in the cytoplasm [17][18][17,18].
2. Zinc Finger Antiviral Protein—ZAP
The zinc finger antiviral protein (ZAP), also known as Poly(ADP-ribose) Polymerase-13 (PARP-13), ADP-Ribosyl-Transferases Diphtheria Toxin-Like-13 (ARTD13), Zinc finger CCCH-type and antiviral 1, is encoded by the human gene ZC3HAV1 (zinc finger CCCH-type containing, antiviral 1, Chromosome 7). ZAP belongs to the PARP protein family (poly(ADP-ribose) polymerase), which uses nicotinamide adenine dinucleotide (NAD+) as a substrate to generate modifications in acceptor proteins but lacks poly(ADP-ribosylation) activity [13][19][20][13,28,29].
ZAP plays a role in the intracellular host cell’s immune system by detecting positive and negative single-stranded RNA viruses from various families during infection. However, it does not bind to double-stranded RNA viruses (dsRNA) [20][29]. ZAP exhibits antiviral properties by directing these viruses to degradation pathways and/or inhibiting their translation, thus restricting viral replication. This prevents the accumulation of viral RNA in the cytoplasm and hinders the virus from multiplying. The viruses affected by ZAP include murine leukemia virus (MLV), Sindbis virus (SINV), and the RNA intermediate of the hepatitis B DNA virus, among others (Table 1) [6][7][13][19][21][22][23][24][6,7,13,28,30,31,32,33].
The presence of orthologs in certain animals, such as mammals, fish, and reptiles, indicates that ZAP has a distant origin [15][25][15,34]. A phylogenetic analysis revealed that the ZAP gene originated in tetrapods [26][35]. Gongalves-Carneiro et al. (2021) [26][35] tested ZAP-related proteins from tetrapods and found that these proteins have an antiviral role in human cells [26][35]. ZAP was initially described in Rat2 cells, where it reduced the replication of MMLV [13] and later in humans [15].
ZAP is consistently expressed in human cells (hZAP) and has two main isoforms derived from alternative splicing. The long isoform, known as ZAP-L or PARP-13.1, consists of 902 amino acids and is associated with the membrane (it contains the YYV catalytic motif). The short isoform, called ZAP-S or PARP13.2, consists of 699 amino acids and is located in the cytosol. Both isoforms originate from the same exon [15][19][27][15,28,36]. Li et al. (2019) [16] discovered two additional splice variants of ZAP in humans: ZAP-XL (extralong) and ZAP-M (medium). However, the ZAP-L and ZAP-S isoforms are the most abundant [15][16][15,16]. Furthermore, they found that the longer isoforms (ZAP-L and ZAP-XL) have a greater impact on alphavirus and hepatitis B virus (HBV) (DNA virus) compared to ZAP-S and ZAP-M [16]. ZAP can be activated by viral infection due to the presence of binding sites for the signal transducer and activator of transcription (STAT) and Interferon Regulatory Factor 3 (IRF3) in its promoter [11][28][29][30][11,37,38,39]. ZAP-L is constitutively expressed in Huh7 cells and acts promptly upon infection, whereas ZAP-S is expressed dependent on IFN signaling [17][18][19][22][31][32][33][17,18,28,31,40,41,42]. Okudera et al. (2022) [34][43] found that treatment of human cells with polyinosinic-polycytidylic acid (poly IC), a Toll-like receptor 3 (TLR3) agonist, upregulated ZAP-S expression. However, when cells were transfected with siRNA against IRF3 or siRNA against TRIM25, the upregulation of ZAP-S was reduced upon treatment with poly-IC [34][43]. The mechanisms through which IRF3 and TRIM25 regulate ZAP-S expression need further clarification.
ZAP in humans has three structural domains: (1) an N-terminal RNA-binding domain (RBD) (amino acids [aa] 1–240) that has four CCCH-type zinc fingers; (2) an integrated central domain (aa 241–700) that contains a TPH domain (TiPARP homology region) containing a fifth zinc finger motif and two WWE modules (Domain in Deltex and TRIP12 homologues (Thyroid Hormone Receptor Interactor 12)); and (3) catalytically inactive C-terminal poly(ADP-ribose) polymerase (PARP)-like domain (aa 701–902) with a regulatory function [15][26][35][36][37][15,25,35,44,45] (Figure 1). According to Kerns et al. (2008) [15], although ZAP-S has a broader expression pattern than ZAP-L, expression of ZAP-L was observed in cell lymphocytes and germline tissues [15].
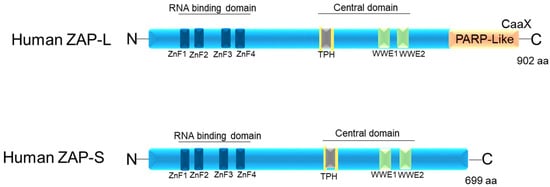
Figure 1. Schematic image of the protein domains of the two isoforms of human ZAP: ZAP-L and ZAP-S. ZnF1–4: four CCCH-type zinc finger motifs. TPH (or TiPARP Homology domain (conserved among ZAP paralogs and containing a fifth CCCH zinc finger motif). WWE motifs. CaaX: C is cysteine, A is usually two aliphatic amino acids, and X can be a variety of amino acids.
The C-terminal poly(ADP-ribose) polymerase (PARP)-like domain is absent in ZAP-S (PARP-13.2 isoform) and enzymatically inactive in ZAP-L, as it lacks the histidine, tyrosine, and glutamate (H-Y-E) catalytic triad [38][46]. However, all isoforms contain both the N-terminal and central domains [13][21][39][40][13,30,47,48] (Figure 1). The targeting of ZAP-L to mem-branes is possible due to the presence of the PARP domain that contains a cysteine (CaaX) motif, which mediates S-farnesylation [18] (Figure 1). This explains its greater presence in vesicular compartments and the cytoplasmic membrane inside the cell, while ZAP-S is mainly found in the cytoplasm [17][31][17,40]. The hypothesis is that ZAP-L can inhibit viruses whose entry into the cell occurs through endocytosis. Furthermore, it may have antiviral activity against viruses that replicate in membrane-derived compartments [41][42][49,50]. Another hypothesis is that ZAP-L is targeted to membranes to form antiviral complexes with its cofactors to exert its antiviral activity. This hypothesis is the most accepted, as ZAP-L targets vRNAs that have several replication sites and mechanisms [15][16][22][37][38][43][44][15,16,31,45,46,51,52].
It is not yet well established whether the two isoforms (ZAP-L and ZAP-S) of ZAP in humans have different roles or overlap in antiviral activity. ZAP-L is known to inhibit alphaviruses and HBV better than ZAP-S [16]. Although both isoforms have the N-terminal domain, they seem to restrict viruses in different ways, as will be discussed later.
A mutation in the cysteine 88 to arginine (ZAPC88R) in the zinc fingers domain of ZAP has been observed to result in the complete loss of antiviral activity [45][53]. The interference of the second (ZnF2) and fourth zinc finger (ZnF4) motifs appears to inhibit the antiviral activity of ZAP, while mutations in other ZnF motifs have a milder impact on the protein’s function [21][46][19,30]. Meagher et al. (2019) [47][54] showed that the ZnF2 motif contains a pocket that selectively packs CG dinucleotides, while ZnF3 contains a binding pocket for guanine and cytosine and ZnF4 for cytosine [46][47][19,54]. ZAP forms a dimer that binds to ZAP-responsive element (ZRE) sequences in the target viral RNA, and these ZREs are specific to ZAP [36][40][48][44,48,55] (Figure 1). Yang et al. (2022) [9] found that mutations affecting the binding of both ZAP and TRIM25 to RNA interfere with antiviral activity against Sindbis virus (SINV). They discovered that mutations in the ZnF of both ZAP-S (mutation in ZnF1 or ZnF3) and ZAP-L (mutation in ZnF1) affected SINV RNA binding. Furthermore, the mutation in ZnF4 of ZAP-L increased SINV replication, but mutations in ZnF from ZAP-S did not, possibly due to the greater antiviral activity of ZAP-L compared to ZAP-S [9].
3. RNA Recognition by ZAP
The target mRNA region to which the ZAP protein binds is rich in CpG (cytosine-phosphate-guanine) dinucleotides, which are two adjacent nucleotides in a linear sequence. After binding, a decrease in vRNA in the cytoplasm is observed, indicating degradation of RNA or translational repression [49][22]. Each ZAP molecule binds to a CpG, allowing it to form an oligomer in the target viral RNA [40][46][47][19,48,54]. This specificity is due to an integrated pocket that accommodates only CpG dinucleotide sequences and is contained within a larger RNA-binding domain [11][14][43][11,14,51].
In vertebrate genomes, the cytosine of a CpG dinucleotide is prone to methylation. This is due to the presence of DNA methyltransferases, enzymes that catalyze the conversion of cytosines in a CpG context to 5-methylcytosine. Methylation is followed by deamination and mutation, resulting in the gradual replacement of CpG dinucleotides by TpG and CpA [50][56]. However, CpGs in RNA viruses are not subject to the same methylation and mutation pressure. Therefore, ZAP has little effect on human mRNA, as vertebrate cells have a suppressed CpG dinucleotide frequency [51][57]. Some viruses have evolved genomes that are suppressed by CpG-enriched segments, avoiding detection by ZAP [49][22]. Based on this, some viruses can modify their CpG dinucleotides in order to develop live attenuated vaccines. Other studies have demonstrated that ZAP detection of foreign genomes also involves RNA secondary structures containing stem loops with conserved sequences, such as “GGGUGG” and “GAGGG” [52][58]. Altering this conserved region leads to diminished RNA recognition and a decrease in ZAP’s antiviral function [52][58]. It is worth noting that ZAP sensitivity to alphaviruses does not correlate with CpG dinucleotide content [16]. Further analysis is needed to determine whether ZAP binding to a specific region of alphavirus RNA is due to structural reasons or linear sequence motifs. Nguyen et al. (2023) [53][59] found that in the genome of ZAP-sensitive alphaviruses (Ross River virus (RRV), Sindbis virus (SINV)), there were three 500 bp sequences correlated with CpG in the non-structural protein (nsP) gene region, compared to ZAP-resistant alphaviruses (o’nyong’nyong virus (ONNV), chikungunya virus (CHIKV)). Furthermore, the nsP2 region of ZAP-sensitive alphaviruses is crucial for ZAP sensitivity, and its binding is CpG-dependent [53][59].
Additionally, viruses with increased CpG dinucleotide content do not always guarantee sensitivity to ZAP. Studies have reported that ZAP is able to restrict viruses with abundant UpA (uracil-phosphate-adenine) dinucleotides [16][37][54][55][56][16,26,27,45,60]. Further research is needed to demonstrate the binding of ZAP to the UpA sequence, the mechanisms behind this binding, and its antiviral role. Gonçalves-Carneiro et al. (2022) [57][61] found that the number, spacing, and surrounding sequence of CpG dinucleotides in the env gene are important for ZAP sensitivity. With this understanding, it is possible to study the generation of a mutant virus genome with modifications that act as a live attenuated vaccine and are precisely prevented by the ZAP protein, as proposed by the author [57][61]. ZAP can bind to RNA with a low frequency of CpG dinucleotides, as the location of these CpG motifs is crucial for these viruses to be restricted [43][44][51,52]. This demonstrates that key points in the understanding of ZAP’s restriction of vRNA still need to be elucidated.
4. Cofactors Required by ZAP for its Antiviral Activity
Given that ZAP lacks RNase activity, it relies on other mechanisms for its antiviral activity. One such mechanism involves its interaction with a cellular polyadenylate-specific ribonuclease (PARN), which degrades the poly(A) tail. ZAP can also recruit an exosomal complex that contains exoribonucleases with 3’-5’ activity, such as ribosomal RNA-processing protein 46/exosome complex component (RRP46/EXOSC5) and ribosomal RNA-processing protein 42/exosome complex component (RRP42/EXOSC7), resulting in the cleavage of viral RNAs [48][58][55,62]. Elements bound to ZAP can also engage the decapping complex (decapping protein 1 (DCP1) and DCP2) through the RNA helicase p72 (DDX17), leading to the removal of the 5’ cap structure of the mRNA. Additionally, the 5’-3’ Exoribonuclease 1 (XRN1) is involved in the process of viral RNA degradation [48][55]. Further studies are needed to investigate the interaction between ZAP isoforms and these proteins with antiviral activities to determine whether there is a preference in the interaction of these proteins with ZAP-L or ZAP-S.
It has also been suggested that ZAP can be observed in cytoplasmic RNA stress granules, even in the absence of viral infection. These stress granules determine the fate of mRNAs that are not involved in translation, either stabilizing them or directing them towards degradation pathways. This suggests that ZAP may be involved in the regulation of cellular mRNA [27][32][36,41].
Since ZAP lacks nuclease activity, it recruits proteins such as TRIM25 to act as cofactors in mediating its antiviral activity [59][60][61][20,21,63]. It is important to identify all the accessory proteins, including those with endonuclease domains, that interact with the N-terminal domain of ZAP to better understand its mechanism of inhibiting viral replication. This highlights the involvement of other host factors that contribute to the innate immune response in addition to the ZAP protein. To gain a better understanding of ZAP’s antiviral activity and its mechanism of action, it is crucial to describe these cofactors in more detail.
4.1. Tripartite Motif Containing 25 (TRIM25)
TRIM25 is a member of the tripartite motif (TRIM) family of proteins, which play a role in supporting the host’s innate immune response to viral infections [62][64]. It is an E3 ubiquitin ligase enzyme that is induced by type I IFN [63][65]. The protein consists of an N-terminal RING domain, followed by a B-box type 1 domain, a B-box type 2 domain, a coiled-coil domain (CCD), and a C-terminal SPRY domain [62][64]. TRIM25 has the ability to bind to RNA, which is crucial for its subcellular localization and antiviral activity [64][66]. Additionally, TRIM25 can modify ZAP-S and ZAP-L through K48- and K63-linked polyubiquitin [59][20].
TRIM25 enhances the antiviral activity of ZAP (Figure 2), but the mechanism behind this cooperation is not well understood. While TRIM25 can bind to both single-stranded and double-stranded RNA [64][66], the involvement of ZAP in this process is not fully understood. ZAP and TRIM25 are both interferon-stimulated genes in human cells [65][67]. According to Li et al. (2017) [59][20], TRIM25 lacking the RING domain or coiled-coil domain loses its ability to stimulate ZAP’s antiviral function [59][20].
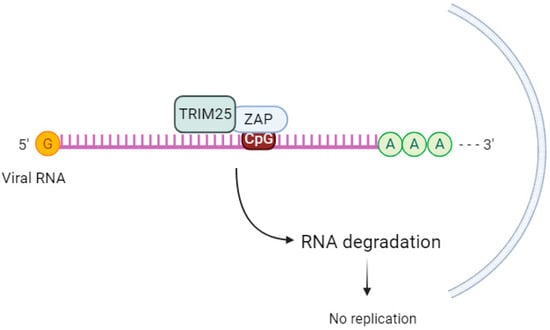
Figure 2. TRIM25 acts as a cofactor of ZAP. ZAP binds to the CpG sequence in viral RNA and, upon sequential binding of TRIM25 and catalytic activation, induces downstream signaling to inhibit viral replication.
The ubiquitination of ZAP by TRIM25 does not seem to be crucial for ZAP’s antiviral activity. When Li et al. (2017) [59][20] used a ZAP-S 7UbΔ mutant that cannot be ubiquitinated, there was still inhibition of SINV replication. This suggests that other factors are involved in TRIM25-mediated upregulation of ZAP’s antiviral activity. The same group discovered that in the absence of TRIM25, ZAP-mediated SINV translation was reduced [59][20]. Other authors [66][68] propose that TRIM25-mediated ubiquitination of other substrates leads to the activation of the antiviral state [66][68].
Gonçalves-Carneiro et al. (2021) [26][35] observed that the functional interaction between TRIM25 and ZAP is inherently protein–protein via the N-terminal ZAP zinc finger domain and the C-terminal SPRY domain of TRIM25, and this interaction is not RNA-dependent. A hypervariable sequence in the N-terminal domain of ZAP was important for the species-specific dependence of the ZAP protein on TRIM25 [26][67][35,69].
In addition to acting as a cofactor of ZAP, TRIM25 can also affect the expression of both ZAP isoforms by regulating alternative splicing, which is necessary for effective ZAP-S expression [68][70]. Yang et al. (2022) [9] demonstrated that the ZAP and TRIM25 interaction acts to inhibit the translation of Japanese encephalitis virus (JEV) in 293T cells transfected with replication-defective JEV replicon RNA reporter and subsequently transfected with a reporter gene to measure luciferase activity [9]. Further studies are needed to clarify the downstream mechanisms after TRIM25 and ZAP interaction, as well as the impact of ZAP ubiquitination on its antiviral activity. Furthermore, studies are needed to verify in what viral contexts TRIM25 regulates ZAP expression.
4.2. KH and NYN Domain Containing (KHNYN)
KHNYN, a cytoplasmic protein containing an NYN ribonuclease domain, has been identified as an accessory protein for the antiviral activity of ZAP. It targets viral RNAs for degradation [69][71]. In cells lacking ZAP but expressing high levels of KHNYN, there was no substantial inhibition of genomic RNA (gRNA) abundance of HIV-1 with CpGs introduced in the Env protein (HIV-1EnvCpG86-561). However, in the absence of KHNYN, viruses multiplied more successfully, demonstrating that KHNYN decreases HIV-1 RNA containing CpG dinucleotides in a ZAP-dependent manner. The authors also observed through the immunoprecipitation assay that KHNYN interacts with ZAP-S and ZAP-L. The KH-like domain and NYN domain endonuclease are required for the antiviral activity of KHNYN. Furthermore, KHNYN decreases Gag and Env expression and virion production. The same group also found that the KHNYN interaction with ZAP is important for the inhibition of HIV-1 containing clustered CpG dinucleotides, and this inhibition requires TRIM25. ZAP and KHNYN can directly interact to form a heterodimer, but TRIM25 is not required for this interaction [69][71] (Figure 3). ZAP targets KHNYN to CpG dinucleotides in viral RNA for cleavage [69][71]. Kmiec et al. (2021) [35][25] demonstrated that the PARP domain containing the CaaX motif in ZAP-L is essential for a more efficient interaction with KHNYN and TRIM25 compared to the ZAP-S interaction with these two cofactors. When ZAP-S received the CaaX box, there was a benefit in the interaction with the cofactors, whereas when the CaaX box was mutated, the viral restriction by ZAP-L was overturned [35][25]. Additionally, they observed that the PARP and CaaX box were essential for antiviral activity against CpG-enriched HIV-1 and SARS-CoV-2. The vesicular localization of ZAP-L seems to correlate with its antiviral action against CpG-enriched HIV-1, and S-farnesylation for ZAP-L was important to inhibit SARS-CoV-2 since this virus replicates in compartments originating from the endoplasmic reticulum (ER) [35][25].
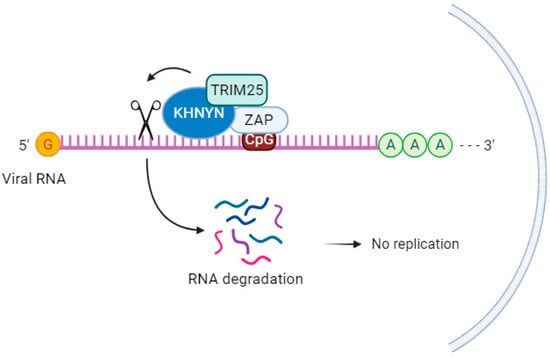
Figure 3. KHNYN acts as a cofactor for the antiviral activity of ZAP. KHNYN interaction with ZAP is important for the inhibition of HIV-1 containing clustered CpG dinucleotides, and this inhibition requires TRIM25.
Further studies are needed to assess how TRIM25 is induced for the interaction between KHNYN and ZAP, the downstream mechanisms after this interaction on viral RNA, and the viral context in which this occurs.
4.3. Exosome
The exosome complex contains a 3’-5’ exoribonuclease activity that contributes to the processing and/or degradation of RNA molecules. It is a multisubunit complex that, in humans, has a core containing nine subunits. This includes six proteins with the PH RNase domain (hRrp41p, hRrp42p, hRrp43p, hRrp46p, PM/Scl-75, and Mtr3), as well as three RNA-binding proteins (Rrp40, Rrp4, and Csl4). There are both nuclear and cytoplasmic forms of the exosome, but it is in the cytoplasm where the degradation of mRNAs containing AU-rich elements (AREs) within their 3’ untranslated regions occurs [58][62]. ZAP targets ZRE-containing mRNAs but not ARE mRNAs [21][30]. When ZAP binds to the ZRE-containing mRNA, it can require the RNA-processing exosome complex [58][62]. The N-terminal domain of human ZAP, which contains the exosome-interacting domains, interacts with the exosome component hRrp42 [70][72]. Inhibition of this component leads to a reduction in the action of ZAP [48][55]. Guo et al. (2007) [58][62] found that ZAP binds to the C-terminal fragment of hRrp46p and that depletion of hRrp46p leads to a decrease in ZAP activity [58][62]. The initiating step of the 3’-5’ decay pathway is the removal of the Poly-A tail by Poly(A) ribonuclease (PARN) [48][55], followed by degradation of the mRNA from the 5’-end by 5’-3’ exoribonucleases (XRNs) or from the 3’-end [71][73]. The interaction between the exosome and ZAP was observed by immunoprecipitation assay [72][73][74,75].
More studies are needed to verify in which context this exosome recruitment occurs by ZAP in the presence of other cofactors.
4.4. p72 RNA Helicase
p72 RNA helicase, also known as p72 DEAD-box RNA helicase or DDX17 [74][75][76,77], is involved in the regulation of RNA structure. It contains a conserved motif Asp-Glu-Ala-Asp (DEAD) and is responsible for ATP-dependent RNA helicase activity. This helicase catalyzes the rearrangement of RNA structure and plays a role in various metabolic processes, including transcription [76][78], translation, RNA degradation [77][79], and pre-mRNA processing/alternative splicing [78][80]. In a study by Chen et al. (2008), it was found that both the N- and C-terminal domains of p72 RNA helicase bind to ZAP, a protein linked to mRNA, in an RNA-independent manner [36][44]. This interaction enhances the efficiency of ZAP in inhibiting virus replication by targeting mRNAs for degradation through the exosome. Additionally, p72 RNA helicase recruits the Dcp1:Dcp2 decapping enzyme to the 5’-end of viral RNAs, inhibiting cap-dependent mRNA translation initiation and inducing viral RNA degradation. It also recruits the complex exoribonuclease XRN1 [48][79][55,81] (Figure 4). Although p72 RNA helicase does not directly interact with the exosome [79][81], it forms an antiviral complex with it when recruited by ZAP. When p72 RNA helicase is depleted using siRNA, ZAP’s antiviral activity is decreased. Further studies are needed to determine which isoforms of ZAP p72 RNA helicase enhance its efficiency against viruses.
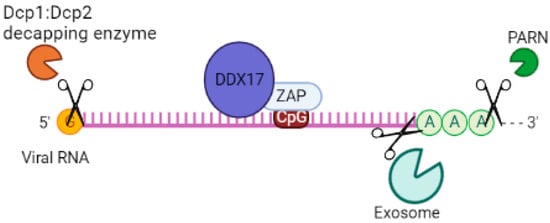
Figure 4. System of cofactors required by ZAP to target viral RNA for degradation. DDX17 increases the efficiency of ZAP by inhibiting virus replication by targeting mRNAs for degradation via the exosome and recruitment of Dcp1:Dcp2 decapping enzyme to the 5′-end of viral RNAs, inhibiting cap-dependent mRNA translation initiation. ZAP interacts with a cellular polyadenylate-specific ribonuclease (PARN), which is a 3’-exoribonuclease that removes poly(A) tails from the 3’ end of RNAs.
References
- Chow, K.T.; Gale, M.; Loo, Y.-M. RIG-I and Other RNA Sensors in Antiviral Immunity. Rev. Immunol.2018, 36, 667–694. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Rev. Virol.2019, 6, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Abernathy, E.; Glaunsinger, B. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology2015, 479-480, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Rev. Immunol.2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, F.; Zhu, Y.; Gao, G. Zinc-finger antiviral protein inhibits XMRV infection. PLoS ONE2012, 7, 39159. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Möller, P.; Bick, M.J.; Wurr, S.; Becker, S.; Günther, S.; Kümmerer, B.M. Inhibition of filovirus replication by the zinc finger antiviral protein. J.2007, 81, 2391–2400. [Google Scholar] [CrossRef] [PubMed]
- Bick, M.J.; Carroll, J.-W.N.; Gao, G.; Goff, S.P.; Rice, C.M.; MacDonald, M.R. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J.2003, 77, 11555–11562. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, X.; Goff, S.P.; Gao, G. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J.2012, 31, 4236–4246. [Google Scholar] [CrossRef]
- Yang, E.; Nguyen, L.P.; Wisherop, C.A.; Kan, R.L.; Li, M.M.H. The Role of ZAP and TRIM25 RNA Binding in Restricting Viral Translation. Cell. Infect. Microbiol.2022, 12, 886929. [Google Scholar] [CrossRef]
- Chiu, H.-P.; Chiu, H.; Yang, C.-F.; Lee, Y.-L.; Chiu, F.-L.; Kuo, H.-C. Inhibition of Japanese encephalitis virus infection by the host zinc-finger antiviral protein. PLOS Pathog.2018, 14, 1007166. [Google Scholar] [CrossRef]
- Wang, N.; Dong, Q.; Li, J.; Jangra, R.K.; Fan, M.; Brasier, A.R.; Lemon, S.M.; Pfeffer, L.M.; Li, K. Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-kappaB-independent. Biol. Chem.2010, 285, 6080–6090. [Google Scholar] [CrossRef] [PubMed]
- Galão, R.P.; Wilson, H.; Schierhorn, K.L.; Debeljak, F.; Bodmer, B.S.; Goldhill, D.; Hoenen, T.; Wilson, S.J.; Swanson, C.M.; Neil, S.J.D. TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction. PLoS Pathog.2022, 18, e1010530. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Guo, X.; Goff, S.P. Inhibition of Retroviral RNA Production by ZAP, a CCCH-Type Zinc Finger Protein. Science2002, 297, 1703–1706. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yan, K.; Wei, L.; Yang, J.; Lu, C.; Xiong, F.; Zheng, C.; Xu, W. Zinc finger antiviral protein inhibits coxsackievirus B3 virus replication and protects against viral myocarditis. Res.2015, 123, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Kerns, J.A.; Emerman, M.; Malik, H.S. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet.2008, 4, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.H.; Aguilar, E.G.; Michailidis, E.; Pabon, J.; Park, P.; Wu, X.; de Jong, Y.P.; Schneider, W.M.; Molina, H.; Rice, C.M.; et al. Characterization of novel splice variants of zinc finger antiviral protein (ZAP). Virol.2019, 93, e00715-19. [Google Scholar] [CrossRef] [PubMed]
- Schwerk, J.; Soveg, F.W.; Ryan, A.P.; Thomas, K.R.; Hatfield, L.D.; Ozarkar, S.; Forero, A.; Kell, A.M.; Roby, J.A.; So, L.; et al. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Immunol.2019, 20, 1610–1620. [Google Scholar] [CrossRef]
- Charron, G.; Li, M.M.; MacDonald, M.R.; Hang, H.C. Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Natl. Acad. Sci. USA2013, 110, 11085–11090. [Google Scholar] [CrossRef]
- Luo, X.; Wang, X.; Gao, Y.; Zhu, J.; Liu, S.; Gao, G.; Gao, P. Molecular Mechanism of RNA Recognition by ZincFinger Antiviral Protein. Cell Rep.2020, 30, 46–52.e4. [Google Scholar] [CrossRef]
- Li, M.M.H.; Lau, Z.; Cheung, P.; Aguilar, E.G.; Schneider, W.M.; Bozzacco, L.; Molina, H.; Buehler, E.; Takaoka, A.; Rice, C.M.; et al. TRIM25 Enhances the Antiviral Action of Zinc-Finger Antiviral Protein (ZAP). PLoS Pathog.2017, 13, 1006145. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, X.; Tu, F.; Wang, Q.; Fan, Z.; Gao, G. TRIM25 Is Required for the Antiviral Activity of Zinc Finger Antiviral Protein. Diamond MS, editor. Virol.2017, 91, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Takata, M.A.; Gonçalves-Carneiro, D.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG dinucleotide suppression enables antiviral defense targeting non-self RNA. Nature2017, 550, 7674. [Google Scholar] [CrossRef] [PubMed]
- Colmant, A.M.G.; Hobson-Peters, J.; Slijkerman, T.A.P.; Harrison, J.J.; Pijlman, G.P.; Van Oers, M.M.; Simmonds, P.; Hall, R.A.; Fros, J.J. Insect-Specific Flavivirus Replication in Mammalian Cells Is Inhibited by Physiological Temperature and the Zinc-Finger Antiviral Protein. Viruses2021, 13, 573. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M.M.; Kibe, A.; Rand, U.; Pekarek, L.; Ye, L.; Buck, S.; Smyth, R.P.; Cicin-Sain, L.; Caliskan, N. The short isoform of the host antiviral protein ZAP acts as an inhibitor of SARS-CoV-2 programmed ribosomal frameshifting. Commun.2021, 12, 7193. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, D.; Lista, M.J.; Ficarelli, M.; Swanson, C.M.; Neil, S.J.D. S-farnesylation is essential for antiviral activity of the long ZAP isoform against RNA viruses with diverse replication strategies. PLoS Pathog.2021, 17, e1009726. [Google Scholar] [CrossRef] [PubMed]
- Fros, J.J.; Visser, I.; Tang, B.; Yan, K.; Nakayama, E.; Visser, T.M.; Koenraadt, C.J.M.; van Oers, M.M.; Pijlman, G.P.; Suhrbier, A.; et al. The dinucleotide composition of the Zika virus genome is shaped by conflicting evolutionary pressures in mammalian hosts and mosquito vectors. PLoS Biol.2021, 19, e3001201. [Google Scholar] [CrossRef]
- Odon, V.; Fros, J.J.; Goonawardane, N.; Dietrich, I.; Ibrahim, A.; Alshaikhahmed, K.; Nguyen, D.; Simmonds, P. The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res.2019, 47, 8061–8083. [Google Scholar] [CrossRef]
- Vyas, S.; Chesarone-Cataldo, M.; Todorova, T.; Huang, Y.H.; Chang, P.A. Systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Commun.2013, 4, 2240. [Google Scholar] [CrossRef]
- Karlberg, T.; Klepsch, M.; Thorsell, A.G.; Andersson, C.D.; Linusson, A.; Schuler, H. Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein. Biol. Chem.2015, 290, 7336–7344. [Google Scholar] [CrossRef]
- Guo, X.; Carroll, J.-W.N.; MacDonald, M.R.; Goff, S.P.; Gao, G. The Zinc Finger Antiviral Protein Directly Binds to Specific Viral mRNAs through the CCCH Zinc Finger Motifs. Virol.2004, 78, 12781–12787. [Google Scholar] [CrossRef]
- Mao, R.; Nie, H.; Cai, D.; Zhang, J.; Liu, H.; Yan, R.; Cuconati, A.; Block, T.M.; Guo, J.-T.; Guo, H. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog.2013, 9, e1003494. [Google Scholar] [CrossRef] [PubMed]
- Todorova, T.; Bock, F.; Chang, P. PARP13 and RNA regulation in immunity and cancer. Trends Mol. Med.2015, 21, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.; Liu, L.; Shen, S.; Deng, H.; Gao, G. Zinc finger antiviral protein inhibits murine gammaherpesvirus 68 M2 expression and regulates viral latency in cultured cells. J.2012, 86, 12431–12434. [Google Scholar] [CrossRef] [PubMed]
- Goossens, K.E.; Karpala, A.J.; Ward, A.; Bean, A.G. Characterisation of chicken ZAP. Comp. Immunol.2014, 46, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves-Carneiro, D.; Takata, M.A.; Ong, H.; Shilton, A.; Bieniasz, P.D. Origin and evolution of the zinc finger antiviral protein. PLoS Pathog.2021, 17, e1009545. [Google Scholar] [CrossRef]
- Leung, A.K.; Vyas, S.; Rood, J.E.R.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Cell2011, 42, 489–499. [Google Scholar] [CrossRef]
- Zhang, Y.; Burke, C.W.; Ryman, K.D.; Klimstra, W.B. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. Virol.2007, 81, 11246–11255. [Google Scholar] [CrossRef]
- MacDonald, M.R.; Machlin, E.S.; Albin, O.R.; Levy, D.E. The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. Virol.2007, 81, 13509–13518. [Google Scholar] [CrossRef]
- Ryman, K.D.; Meier, K.C.; Nangle, E.M.; Ragsdale, S.L.; Korneeva, N.L.; Rhoads, R.E.; MacDonald, M.R.; Klimstra, W.B. Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells. Virol.2005, 79, 1487–1499. [Google Scholar] [CrossRef]
- Hayakawa, S.; Shiratori, S.; Yamato, H.; Kameyama, T.; Kitatsuji, C.; Kashigi, F.; Goto, S.; Kameoka, S.; Fujikura, D.; Yamada, T.; et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Immunol.2011, 12, 37–44. [Google Scholar] [CrossRef]
- Seo, G.J.; Kincaid, R.P.; Phanaksri, T.; Burke, J.M.; Pare, J.M.; Cox, J.E.; Hsiang, T.-Y.; Krug, R.M.; Sullivan, C.S. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe2013, 14, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Todorova, T.; Bock, F.J.; Chang, P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 Transcript. Commun.2014, 5, 5362. [Google Scholar] [CrossRef] [PubMed]
- Okudera, M.; Odawara, M.; Arakawa, M.; Kawaguchi, S.; Seya, K.; Matsumiya, T.; Sato, R.; Ding, J.; Morita, E.; Imaizumi, T. Expression of Zinc-Finger Antiviral Protein in hCMEC/D3 Human Cerebral Microvascular Endothelial Cells: Effect of a Toll-Like Receptor 3 Agonist. Neuroimmunomodulation2022, 29, 349–358. [Google Scholar] [CrossRef]
- Zhu, Y.; Gao, G. ZAP-mediated mRNA degradation. RNA Biol.2008, 5, 65–67. [Google Scholar] [CrossRef]
- Gonzalez-Perez, A.C.; Stempel, M.; Wyler, E.; Urban, C.; Piras, A.; Hennig, T.; Ganskih, S.; Wei, Y.; Heim, A.; Landthaler, M.; et al. The Zinc Finger Antiviral Protein ZAP Restricts Human Cytomegalovirus and Selectively Binds and Destabilizes Viral UL4/UL5 Transcripts. mBio2021, 12, e02683-20. [Google Scholar] [CrossRef] [PubMed]
- Goodier, J.L.; Pereira, G.C.; Cheung, L.E.; Rose, R.J.; Kazazian, H.H.J. The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition. PLoS Genet.2015, 11, e1005252. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Braczyk, K.; Gonçalves-Carneiro, D.; Dawidziak, D.M.; Sanchez, K.; Ong, H.; Wan, Y.; Zadrozny, K.K.; Ganser-Pornillos, B.K.; Bieniasz, P.D.; et al. Poly(ADP-ribose) potentiates ZAP antiviral activity. PLoS Pathog.2022, 18, e1009202. [Google Scholar] [CrossRef]
- Chen, S.; Xu, Y.; Zhang, K.; Wang, X.; Sun, J.; Gao, G.; Liu, Y. Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Struct. Mol. Biol.2012, 19, 430–435. [Google Scholar] [CrossRef]
- Soveg, F.W.; Schwerk, J.; Gokhale, N.S.; Cerosaletti, K.; Smith, J.R.; Pairo-Castineira, E.; Kell, A.M.; Forero, A.; Zaver, S.A.; Esser-Nobis, K.; et al. Endomembrane targeting of human OAS1 p46 augments antiviral activity. eLife2021, 10, e71047. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Sugrue, E.; Lytras, S.; Kuchi, S.; Noerenberg, M.; Turnbull, M.L.; Loney, C.; Herder, V.; Allan, J.; Jarmson, I.; et al. A Prenylated dsRNA Sensor Protects Against Severe COVID-19. Science2021, 374, eabj3624. [Google Scholar] [CrossRef]
- Nchioua, R.; Kmiec, D.; Muller, J.A.; Conzelmann, C.; Gross, R.; Swanson, C.M.; Neil, S.J.D.; Stenger, S.; Sauter, D.; Münch, J.; et al. SARS-CoV-2 is restricted by zinc finger antiviral protein despite preadaptation to the low-CpG environment in humans. mBio2020, 11, e01930-20. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, D.; Nchioua, R.; Sherrill-Mix, S.; Sturzel, C.M.; Heusinger, E.; Braun, E.; Gondim, M.V.P.; Hotter, D.; Sparrer, K.M.J.; Hahn, B.H.; et al. CpG Frequency in the 5′ Third of the env Gene Determines Sensitivity of Primary HIV-1 Strains to the Zinc-Finger Antiviral Protein. mBio2020, 11, e02903-19. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.M.J.; Albin, O.R.; Carroll, J.W.N.; Jones, C.T.; Rice, C.M.; MacDonald, M.R. Identification of a dominant negative inhibitor of human zinc finger antiviral protein reveals a functional endogenous pool and critical homotypic interactions. Virol.2010, 84, 4504–4512. [Google Scholar]
- Meagher, J.L.; Takata, M.; Gonçalves-Carneiro, D.; Keane, S.C.; Rebendenne, A.; Ong, H.; Orr, V.K.; MacDonald, M.R.; Stuckey, J.A.; Bieniasz, P.D.; et al. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CGrich viral sequences. Natl. Acad. Sci. USA2019, 116, 24303–24309. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chen, G.; Lv, F.; Wang, X.; Ji, X.; Xu, Y.; Sun, J.; Wu, L.; Zheng, Y.-T.; Gao, G. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Natl. Acad. Sci. USA2011, 108, 15834–15839. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.N.; Gerber-Huber, S. DNA methylation and CpG suppression. Differ.1985, 17, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Rev. Genet.2010, 11, 204–220. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, X.; Gao, G. Analyses of SELEX-derived ZAP-binding RNA aptamers suggest that the binding specificity is determined by both structure and sequence of the RNA. Protein Cell2010, 1, 752–759. [Google Scholar] [CrossRef]
- Nguyen, L.P.; Aldana, K.S.; Yang, E.; Yao, Z.; Li, M.M.H. Alphavirus Evasion of Zinc Finger Antiviral Protein (ZAP) Correlates with CpG Suppression in a Specific Viral nsP2 Gene Sequence. Viruses2023, 15, 830. [Google Scholar] [CrossRef]
- Goonawardane, N.; Nguyen, D.; Simmonds, P. Association of Zinc Finger Antiviral Protein Binding to Viral Genomic RNA with Attenuation of Replication of Echovirus 7. mSphere2021, 6, e01138-20. [Google Scholar] [CrossRef]
- Gonçalves-Carneiro, D.; Mastrocola, E.; Lei, X.; DaSilva, J.; Chan, Y.F.; Bieniasz, P.D. Rational attenuation of RNA viruses with zinc finger antiviral protein. Microbiol.2022, 7, 1558–1567. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, J.; Sun, J.; Gao, G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. PNAS2007, 104, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lou, D.I.; Sawyer, S.L. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet.2011, 7, e1002388. [Google Scholar] [CrossRef] [PubMed]
- Martín-Vicente, M.; Medrano, L.M.; Resino, S.; García-Sastre, A.; Martínez, I. TRIM25 in the Regulation of the Antiviral Innate Immunity. Immunol.2017, 22, 1187. [Google Scholar] [CrossRef] [PubMed]
- Carthagena, L.; Bergamaschi, A.; Luna, J.M.; David, A.; Uchil, P.D.; Margottin-Goguet, F.; Mothes, W.; Hazan, U.; Transy, C.; Pancino, G.; et al. Human TRIM gene expression in response to interferons. PLoS ONE2009, 4, e4894. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.G.; Sparrer, K.M.J.; Chiang, C.; Reis, R.A.; Chiang, J.J.; Zurenski, M.A.; Wan, Y.; Gack, M.U.; Pornillos, O. TRIM25 Binds RNA to Modulate Cellular Anti-viral Defense. Mol. Biol.2018, 430, 5280–5293. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.E.; Hughes, J.; Gu, Q.; Behdenna, A.; Singer, J.B.; Dennis, T.; Orton, R.J.; Varela, M.; Gifford, R.J.; Wilson, S.J.; et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol.2017, 15, e2004086. [Google Scholar] [CrossRef]
- Yang, E.; Huang, S.; Jami-Alahmadi, Y.; McInerney, G.M.; Wohlschlegel, J.A.; Li, M.M.H. Elucidation of TRIM25 ubiquitination targets involved in diverse cellular and antiviral processes. PLoS Pathog.2022, 18, e1010743. [Google Scholar] [CrossRef]
- Choudhury, N.R.; Heikel, G.; Trubitsyna, M.; Kubik, P.; Nowak, J.S.; Webb, S.; Granneman, S.; Spanos, C.; Rappsilber, J.; Castello, A.; et al. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol.2017, 15, 10. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Chiweshe, S.; McCormick, D.; Raper, A.; Wickenhagen, A.; DeFillipis, V.; Gaunt, E.; Simmonds, P.; Wilson, S.J.; Grey, F. Human cytomegalovirus evades ZAP detection by suppressing CpG dinucleotides in the major immediate early 1 gene. PLoS Pathog.2020, 16, e1008844. [Google Scholar] [CrossRef]
- Ficarelli, M.; Wilson, H.; Pedro Galão, R.; Mazzon, M.; Antzin-Anduetza, I.; Marsh, M.; Neil, S.J.; Swanson, C.M. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. eLife2019, 8, e46767. [Google Scholar] [CrossRef] [PubMed]
- Malgras, M.; Garcia, M.; Jousselin, C.; Bodet, C.; Lévêque, N. The Antiviral Activities of Poly-ADP-Ribose Polymerases. Viruses2021, 13, 582. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Jensen, T.H. The exosome: A multipurpose RNA-decay machine. Trends Biochem. Sci.2008, 33, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Brodersen, D.E.; Jensen, T.H. Origins and activities of the eukaryotic exosome. Cell Sci.2009, 122, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Chlebowski, A.; Lubas, M.; Jensen, T.H.; Dziembowski, A. RNA decay machines: The exosome. Biophys. Acta Gene Regul. Mech.2013, 1829, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Cordin, O.; Banroques, J.; Tanner, N.K.; Linder, P. The DEAD-box protein family of RNA helicases. Gene2006, 367, 17–37. [Google Scholar] [CrossRef]
- Linder, P.; Jankowsky, E. From unwinding to clamping: The DEAD box RNA helicase family. Rev. Mol. Cell Biol.2011, 12, 505–516. [Google Scholar] [CrossRef]
- Fuller-Pace, F.V. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res.2006, 34, 4206–4215. [Google Scholar] [CrossRef]
- Wortham, N.C.; Ahamed, E.; Nicol, S.M.; Thomas, R.S.; Periyasamy, M.; Jiang, J.; Ochocka, A.M.; Shousha, S.; Huson, L.; Bray, S.E.; et al. The DEAD-box protein p72 regulates ERa-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERa-positive breast cancer. Oncogene2009, 28, 4053–4064. [Google Scholar] [CrossRef]
- Honig, A.; Auboeuf, D.; Parker, M.M.; O’Malley, B.W.; Berget, S.M. Regulation of alternative splicing by the ATP-dependent DEADbox RNA helicase p72. Cell. Biol.2002, 22, 5698–5707. [Google Scholar] [CrossRef]
- Chen, G.; Guo, X.; Lv, F.; Xu, Y.; Gao, G. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Natl. Acad. Sci. USA2008, 105, 4352–4357. [Google Scholar] [CrossRef]
