Point-of-care (POC) technologies, which offer immediate and accurate testing at or near the site of patient care, have become a cornerstone of modern medicine. Prokaryotic Argonaute proteins (pAgo), proficient in recognizing target RNA or DNA with complementary sequences, have emerged as potential game-changers. pAgo present several advantages over the currently popular CRISPR/Cas systems-based POC diagnostics, including the absence of a protospacer adjacent motif (PAM) sequence requirement, the use of shorter nucleic acid molecules as guides, and a smaller protein size.
- viral detection
- point-of-care diagnostics
- prokaryotic argonaute
1. Introduction
2. Argonaute Proteins: Structure and Function
2.1. Structural Overview
Argonaute proteins, a diverse family of nucleic acid-guided proteins, are ubiquitously found across various organisms [5][8]. Despite the low sequence homology between eAgo and pAgo, their fundamental structure and function remain highly conserved [6][7][13,14]. eAgo plays a crucial role in the RNA interference (RNAi) process, forming RNA-induced silencing complexes, binding to guide small single-stranded RNA molecules, and leveraging its inherent nuclease activity to either directly cleave target RNA or recruit other silencing proteins for target transcriptional repression [8][15]. pAgo, initially discovered in bacteria and archaea, can be classified into three structural categories: long pAgo, short pAgo, and PIWI-RE proteins [9][16]. Long pAgo is a well-studied class of Ago proteins, featuring structural domains highly reminiscent of eAgo, including N-terminal, PAZ (Piwi-Argonaute-Zwille), MID (middle), and PIWI (P-element Induced Wimpy Testis) domains (Figure 1A). The overall structure forms a bilobed scaffold, with one lobe comprising the N-PAZ domains and the other the MID-PIWI domains [8][15] (Figure 1A). The N-terminal domain, being the least conserved, may facilitate the separation of RNA duplexes. The PAZ domain, a small domain of approximately 140 amino acids, is involved in binding the 3′ end of the guide molecule. The MID domain houses a pocket for binding to the 5′ end of the guide. Upon target binding, the catalytically active PIWI domain mediates target strand cleavage. This domain contains a conserved tetra-amino acid residue DEDX (where X is an aspartate, histidine, or lysine residue), which can bind to the two divalent cations required for catalytic activity [8][15].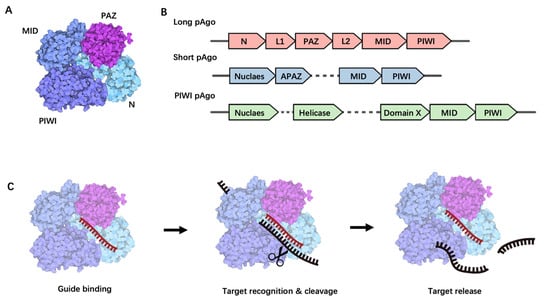
2.2. Biological Functions of Long pAgo
pAgo proteins are hypothesized to play a significant role in cellular defense against viral invasion, a notion supported by recent studies on long pAgo proteins. Firstly, bioinformatics research reveals that Argonaute genes often co-locate in the same operon as genes encoding host defense proteins, such as restriction endonucleases [11][19]. Secondly, some prokaryotic Argonautes contain Silent Information Regulator 2 (SIR2) or Toll/interleukin-1 receptor (TIR) domains, either within the protein or in neighboring genes [10][17]. These domains are typically part of the bacterial anti-phage Thoeris defense mechanism [12][13][20,21]. TtAgo, derived from Thermus thermophilus [14][25], reduces plasmid transformation efficiency and is presumed to serve as a defense against foreign nucleic acid invasion. Further research shows that TtAgo mediates DNA-guided DNA interference, directly cleaving and invading foreign nucleic acids in bacteria at the DNA level [15][26]. In vivo, TtAgo forms a complex with small interference DNA (siDNA) guide molecules (13–25 nucleotides in length) and directs single-stranded target DNA binding and cleavage. In some cases, two complementary TtAgo-siDNA complexes can introduce dsDNA breaks [14][25] via which TtAgo degrades invading DNA and reduces the levels of plasmids in cells. In vitro, TtAgo cleaves the phosphodiester bond between the 10th and 11th bases of the target DNA from the 5′ end using a 5′-phosphorylated ssDNA as the guide DNA (gDNA). PfAgo, derived from Pyrococcus furiosus, exhibits a higher reaction temperature range (87 °C to 99.9 °C) compared to TtAgo. Similar to other bacterial-derived pAgos, the archaeal PfAgo protein reduces the plasmid transformation efficiency of Pyrococcus furiosus, suggesting a role for PfAgo in defending against foreign nucleic acid invasion [16][22]. Studies on PfAgo’s in vitro cleavage activity reveal that PfAgo can bind to 5′-phosphorylated ssDNA (gDNA) at high temperatures and direct the cleavage of complementary single-stranded DNA. The results suggest that gDNA within the length range of 15–31 nt better mediates PfAgo’s cleavage of single-stranded DNA (ssDNA). Other long pAgos include AfAgo (from Archaeoglobus fulgidus), AaAgo (from Aquifex aeolicus), MpAgo (from Marinitoga piezophila), and TpAgo (from Thermotoga profunda). In vitro, AfAgo prefers ssDNA as the guide nucleic acid sequence to bind with target DNA, whereas AaAgo is involved in DNA-mediated RNA cleavage [11][19]. Genes encoding MpAgo and TpAgo are often found in the same gene cluster as those encoding CRISPR-Cas enzymes, suggesting functional links between the two [17][28]. Unlike most pAgos that utilize 5′-phosphorylated gDNA, MpAgo and TpAgo use 5′-hydroxylated gRNA as guides to cleave specific ssDNA [17][28]. Moreover, MpAgo can also use 5′-hydroxylated RNA guides to cleave target ssRNA [17][28], suggesting that although the roles of MpAgo and TpAgo may be similar to other long pAgos, their mechanisms for generating and binding guide molecules are different [17][28].3. pAgo-Based POC Detection Technologies
The utilization of pAgos for viral nucleic acid detection encompasses a series of steps: Initially, viral nucleic acid was extracted from clinical samples and the target (either DNA or RNA) undergoes amplification via PCR or isothermal amplification, with RNA viruses necessitating an extra reverse transcription step (Figure 2A,B). Following this, a 5′ phosphorylated guide nucleic acid is designed to target the sequence, directing pAGO to execute the first round of cleavage on the target DNA amplicon (Figure 2C–E). This process yields secondary 5′ phosphorylated guide nucleic acids originating from the target sequence. These 5′ phosphorylated terminal DNA fragments act as new guide strands to cleave fluorophore-quencher labeled probes (Figure 2F), which can subsequently be detected via fluorescence spectrophotometry, quantitative real-time PCR (qPCR), droplet digital PCR (ddPCR), or a microplate reader (Figure 2H,I). Notably, pAGO, devoid of trans-cleavage activity, can cleave multiple targets and corresponding probes by designing several guide strands, thereby facilitating multiplex targeting of viruses [18][19][20][40,41,42].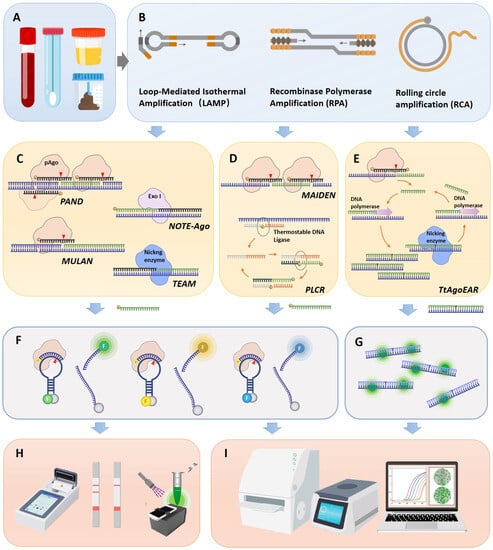
3.1. PfAgo-Mediated Nucleic Acid Detection, PAND
3.2. Ultra-Short PCR and Pyrococcus furiosus Argonaute Combined Nucleic Acid Detection (USPCRP)
He et al. devised a USPCRP nucleic acid detection system that amalgamates ultra-short PCR (usPCR) amplification technology with the PfAgo cleavage system [22][48]. This approach does not necessitate input gDNA but rather employs two primers shorter than 14 nt (one modified with a 5′ phosphate group) to amplify target DNA via ultra-short PCR. This results in a DNA product with a 5′ phosphate group modification, which serves as gDNA for the directed cleavage of fluorescent probes. Given that PfAgo is insensitive to gDNA shorter than 14 nt, the amplification primers with 5′ phosphate group modification do not induce nonspecific cleavage of the fluorescent probes. The USPCRP system was utilized to detect the ORF1ab gene of SARS-CoV-2, MERS-CoV, and SARS-CoV, demonstrating high sensitivity (10 aM) and high specificity single-base resolution nucleic acid detection, and successfully identifying the virus in clinical samples acquired from nasopharyngeal or oropharyngeal swabs, exhibiting a high level of concordance with the outcomes obtained via RT-qPCR. The system generates functional gDNA using usPCR, significantly reducing target fragment enrichment time. The paramount innovation of this system, when contrasted with analogous technologies, resides in its freedom from PAM constraints, necessitating merely two enzymes (Taq DNA polymerase and PfAgo), and its amplification of the detection system’s operability.3.3. Ago-Directed Specific Target Enrichment and Detection (A-Star)
A-Star is an ultra-sensitive, single-tube, single-nucleotide mutation enrichment detection system [23][49]. This system incorporates mismatched and phosphorylated gDNA into the PCR reaction system, directing PfAgo to specifically cleave wild-type sequences during the denaturation step of PCR. This process rapidly enriches rare mutant alleles in the subsequent amplification step, with the amplified products then being suitable for various downstream detection methods, including Sanger sequencing and TaqMan quantitative real-time PCR. Compared to traditional methods, A-Star significantly enhances the sensitivity of detecting rare variant alleles, capable of specifically amplifying Single nucleotide variants (SNV) alleles as low as 0.01%, with an enrichment efficiency exceeding 5500 times.3.4. Multiplex Ago-Based Nucleic Acid Detection System (MULAN)
Ye et al. developed a multi-target detection platform, MULAN, by integrating RT-LAMP isothermal amplification technology with the PfAgo cleavage system [24][43]. This platform consolidates the nucleic acid detection process into a single-tube system, facilitating multiplex nucleic acid detection of different fluorophores via portable devices or qPCR instruments. Using a meticulous design of gDNA and reporter probes, the authors employed this novel method to swiftly identify SARS-CoV-2 WT and its D614G variant. The detection time for RT-LAMP is 35 min, and PfAgo cleavage takes 15 min, with a limit of detection (LoD) for each reaction as low as five copies, surpassing the Cas13a-based SHERLOCK detection system (LoD of 42 copies per reaction within 60 min). Clinically validated, MULAN is capable of swiftly conducting triplex detection of SARS-CoV-2, influenza A, and influenza B viruses in one reaction. This was achieved using nasopharyngeal swabs from patients with confirmed influenza viruses and Pseudoviruses representing SARS-CoV-2.3.5. TtAgo-Assisted Exponential Isothermal Amplification for Multiplex Detection (TEAM)
TEAM is a multiplex nucleic acid detection system based on TtAgo that merges programmable TtAgo cleavage with exponential amplification reaction (EXPAR) for multiplex detection [19][41]. While traditional EXPAR provides excellent amplification efficiency, its sensitivity is limited due to non-specific background signals. By combining Ago’s programmability, precise specificity, and multi-round cleavage activity with EXPAR’s efficient amplification, TEAM achieves single nucleotide discrimination and high sensitivity down to single-molecule concentrations. TtAgo significantly accelerates the detection process, reporting the presence of miRNA in just 10–15 min, reducing the total time for miRNA detection to approximately 30–35 min. By incorporating different fluorescently labeled probes into the detection system, the TEAM system supports the simultaneous detection of four groups of target nucleic acids. The performance of the TEAM assay in diagnosing colorectal cancer (CRC) was evaluated by concurrently detecting several circulating miRNAs in clinical serum samples.3.6. Mesophilic Ago-Based Isothermal Detection Method (MAIDEN)
Li et al. ingeniously tackled the challenge of the high-temperature requirement inherent in TtAgo or PfAgo-based methods [25][29]. They achieved this by broadening their research scope to include mesophilic Agos, thereby facilitating isothermal detection of the CRISPR-like mechanism. This novel approach, termed Mesophilic Ago-Based Isothermal Detection Method (MAIDEN), integrates mesophilic Ago cleavage with reverse transcription. This combination yields single-strand DNA as a substrate and facilitates the cleavage of fluorescence probes, enabling the detection of in vitro transcribed SARS-CoV-2 RNA at moderate temperatures. The initial phase involved mining and optimizing the mesophilic Ago and the fluorescence reporter system, followed by identifying a compatible reverse transcription reaction.3.7. Tt Argonaute-Based Thermostable Exponential Amplification Reaction (TtAgoEAR)
In an effort to consolidate the pAgo-mediated cleavage step and the amplification step into a singular isothermal reaction for RNA analysis, Yuan et al. developed an innovative isothermal amplification strategy. This approach, named Thermus thermophilus Argonaute-based Thermostable Exponential Amplification Reaction (TtAgoEAR) [26][44], enables RNA detection with ultra-sensitivity and single-nucleotide resolution at a constant temperature of 66 °C. The TtAgoEAR system operates using two sequential circuits. The first circuit is designed to derive the desired single-stranded initial oligonucleotide from an RNA target. A 16-nt gDNA, complementary to the target RNA and phosphorylated at both the 5′ and 3′ ends, is designed to activate Ago proteins while inhibiting its extension. The gDNA and TtAgo form a complex that specifically recognizes RNA and generates a nick, leading to the formation of the initial oligonucleotide. The second circuit’s objective is to amplify the initial oligonucleotide released from the first circuit using a single template. This template comprises a central nicking enzyme recognition site and a repetitive sequence complementary to the trigger at both ends. Upon the addition of the target RNA, the produced initial oligonucleotide hybridizes to the template and is extended to form double-stranded DNA by a DNA polymerase. The cleavage of double-stranded DNA by a nicking enzyme generates a new trigger sequence that initiates the repeated cycle of DNA replication via a cycle of hybridization, elongation, and cutting reactions. The exponential reaction process of TtAgoEAR can be monitored in real time using the dye SYBR Green I. TtAgoEAR enables the detection of different types of RNA, showcasing the effectiveness of the coupled TtAgo cleavage and thermostable EXPAR in the TtAgoEAR method.3.8. Short Prokaryotic Argonaute/TIR-APAZ (SPARTA)-Based Nucleic Acid Detection Tool
Koopal B and colleagues delineated the functionality and mechanism of a novel short prokaryotic Argonaute/TIR-APAZ (SPARTA) defense system [27][50]. The SPARTA systems are structured with a catalytically inactive short pAgo and a TIR-APAZ protein, together forming a heterodimeric complex. In this arrangement, the short pAgo operates as the ‘sensor,’ employing guide RNAs to attach to single-stranded (ss) DNA targets. Upon the binding of a target, the NAD(P)ase activity of the TIR-APAZ ‘effector’ becomes unleashed. In vivo, SPARTA targets invading plasmid DNA, causing a reduction in the cellular levels of metabolites NAD+ and NADP+, subsequently leading to cell death. In the application of SPARTA for ssDNA detection, synthetic RNA guides complementary to ssDNA, along with ɛ-NAD (an analogue of NAD+), are introduced into the system. When the target ssDNA is present, SPARTA is activated to form a complex, which subsequently converts ɛ-NAD into fluorescent ɛ-ADPR. SPARTA can also be modified to detect dsDNA, enhancing the sensitivity of SPARTA-based detection. Target DNA can be specifically amplified via PCR using a phosphorothioate (PT) forward primer and an unmodified reverse primer. After incubation with T7 exonuclease, the unmodified strand is degraded, leaving behind ssDNA fragments containing the target sequence detectable by SPARTA. While the detection methods based on SPARTA are still in the conceptual phase, they pave the way for potential point-of-care (POCT) applications of this technology.4. Conclusions
Argonaute (Ago)-based nucleic acid biosensors, when juxtaposed with traditional detection techniques, offer a multitude of advantages, thereby positioning them as a promising platform for the next generation of nucleic acid biosensing. One of the key advantages of Argonaute is its lack of a PAM sequence requirement in target DNA, which provides greater flexibility in the selection of nucleic acid targets. Furthermore, the guide molecules of the majority of prokaryotic Argonautes are short DNA molecules [9][16]. Their high stability, simplicity, and lower production costs make them ideal for the creation of cost-effective, easy-to-store reagents for point-of-care testing (POCT) applications, thereby streamlining both laboratory and field-testing procedures. The high specificity between guide DNA (gDNA) or guide RNA (gRNA) and target nucleic acids facilitates the detection of single-base mutations in viruses. Additionally, compared to other in situ detection technologies, Ago-based biosensors are more adept at achieving multiplex nucleic acid detection [18][28][29][37,40,57].
