Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 1 by Antonio Sasso.
Surface-enhanced Raman scattering (SERS) is of growing interest for a wide range of applications, especially for biomedical analysis, thanks to its sensitivity, specificity, and multiplexing capabilities. A crucial role for successful applications of SERS is played by the development of reproducible, efficient, and facile procedures for the fabrication of metal nanostructures (SERS substrates). Even more challenging is to extend the fabrication techniques of plasmonic nano-textures to atomic force microscope (AFM) probes to carry out tip-enhanced Raman spectroscopy (TERS) experiments, in which spatial resolution below the diffraction limit is added to the peculiarities of SERS.
- Raman spectroscopy
- surface- and tip-enhanced Raman scattering
- biophotonics
1. Introduction
Living systems are highly complex systems, which makes them challenging for quantitative investigations. Approaches in current biology and biotechnology research are increasingly aimed at the identification and precise characterization of basic processes on the level of individual cells or even biomolecules, like proteins, amino acids, and lipids.
Biophotonics is an emerging interdisciplinary research area, born from the merging of biology and photonics. Its purpose is to generate and handle light (photons) to image, detect, and manipulate biological materials [1,2,3,4][1][2][3][4]. The great advantage of using light derives from its non-invasiveness and from the numerous effects arising from light–matter interactions which allow researchers to probe the properties of bio-systems over a large spatial scale, ranging from organs or tissues down to single biomolecules. Similarly, in the time domain, basic molecular mechanisms can be studied over time down to the femtosecond scale. Photonic-based approaches have recently allowed researchers to shed light on the peculiar function of single proteins, DNA, and other important molecules. In medicine, biophotonics has introduced new ways to image and analyze living microorganisms, and for the diagnosis and treatment of diseases [5].
Confocal laser-based fluorescence microscopy is one of the most popular tools for the optical imaging of biomaterials, thanks to its high sensitivity and facile preparation of samples (staining). Generally, fluorescence imaging is achieved by using the intrinsic fluorescence of certain proteins or, more frequently, by resorting to the use of fluorophores bound to target molecules. Fluorescence provides information about the average life of excited states, quantum efficiency, and the degree of depolarization. Such parameters generally change depending on the fluorophore environment (polarity, pH, temperature, etc.) and, therefore, can be used as probes for biological sensors. On the other hand, all fluorescence-based techniques present several shortcomings: for instance, the presence of fluorophores can interfere with the normal behavior of the biomolecule and, most importantly, the chemical selectivity of fluorescence spectra is quite poor. Further, phenomena such as photo-quenching and photo-bleaching, in general, limit the observation time and, hence, the signal-to-noise ratio of the measurements [6,7,8][6][7][8].
Over the past decades, Raman spectroscopy (RS) has attracted much attention for its ability to overcome many of the limitations of fluorescence spectroscopy. Since the pioneering studies of the Indian physicist C.V. Raman (1928) [9], the Raman effect has remained for many decades a poorly investigated phenomenon due to the objective experimental difficulties related to the weak signals originating from inelastic photons. Thanks to the development of laser sources and low noise detectors, RS has recently become a widely used technique for many studies in physics, chemistry, materials science, environmental science, and, especially life science [10,11,12,13,14,15,16][10][11][12][13][14][15][16].
2. Basic Principles of Raman Spectroscopy
As is well known, photons of frequency νinc can be scattered by molecules elastically or inelastically: the former are assigned to Rayleigh scattering, while the shifted photons are associated with Raman scattering. In the context of a quantum description, Rayleigh photons derive from the excitation of a virtual energy level followed by an instantaneous decay towards the same starting vibrational level (νRay = νinc). Conversely, Raman photons (νRaman ≠ νinc) can arise from two different decay channels. If the molecule occupies the lowest vibrational energy level, the decay occurs towards the first excited vibrational level, and the emitted photons exhibit an energy lower than the incident photons (Stokes photons, νStokes < νinc). On the other hand, when the molecule is initially on an excited vibrational level, the decay can take place towards the fundamental level and the emitted photons show an energy higher than the incident photons (anti-Stokes photons, νanti-Stokes > νinc). In both cases, the Raman signal involves a coupling to the internal degree of freedom of the molecule, i.e., the molecular vibration of frequency νvib. In particular, Raman photons have frequency νRaman = νinc ± νvib, where the sum/difference results in anti-Stokes/Stokes Raman scattering, respectively. According to Boltzmann’s statistics, at room temperature, the vibrational levels are poorly occupied; therefore, the anti-Stokes signal is much less intense than the Stokes one. Importantly, since the shift in energy of Stokes photons is associated with the discrete vibrational modes of polarizable molecules, Raman spectra represent an analytical tool for determining the biochemical composition of the investigated analyte (“chemical fingerprint”). In this regard, RS provides information quite similar to that deriving from IR absorption. However, the two techniques have experienced a quite different fortune for biological applications. This is mainly due to the strong IR-absorption cross section of water, which usually masks the contribution of species in aqueous environment. In contrast, water interferes only poorly with Raman spectrum of aqueous solutions, due to the quite low water Raman activity. Focusing on the study of biological material, the most significant spectral regions fall within 400–2000 cm−1 wavenumbers, typically associated with the bond vibrations of relevant macromolecules. In particular, Amide I-III protein bands fall in this range and provide useful insight on protein secondary structures; carbohydrates lie in the 1500–1700 cm−1 spectral range, while phosphate groups of DNA can be found in the 470–1200 cm−1 region. Higher-frequency bond vibrations associated with CH, NH, and OH stretching in lipids and proteins are instead observed in the range 2700–3500 cm−1 [24][17]. However, due to the high number of vibrational degrees of freedom that occurs in complex molecules made up of numerous atoms, a typical Raman spectrum of biological material is overly complex, and the extraction of biological information is quite challenging. Therefore, multivariate analysis approaches are often necessary to distinguish the different molecular components present in the specimen. The other side of the coin of Raman analysis is related to the exceptionally low probability of occurrence of Raman scattering: each Raman photon corresponds to ~106 Rayleigh scattered photons. The strength of the Raman effect can also increase by some order of magnitude if the laser frequency is resonant with molecular electronic transitions (resonance Raman scattering, RRS), but in such conditions, the contrast of Raman peaks could be reduced by the co-presence of a broad fluorescence spectrum. Such negative features of Raman spectroscopy have relegated this technique to specific and rare applications for decades after its discovery. Its recent impressive diffusion is to be found in the extraordinary technological progress related to efficient laser sources, low-noise solid-state detectors, and effective filters to remove Rayleigh background. From an experimental point of view, a Raman spectrum is recorded through the following steps: collecting the scattered photons, rejecting the intense Rayleigh photons (usually by a notch-filter), dispersing the inelastic photons by means of efficient diffraction gratings, and recording the peaks with low-noise CCD camera [25][18]. In recent times, Raman spectroscopy has been combined with confocal optical microscopy (micro-Raman spectroscopy) and even with optical tweezers (Raman tweezers) [26,27][19][20] to analyze single microparticles, bacteria, or cells [28,29][21][22]. The spatial resolution of a micro-Raman system is limited by the diffraction of light and, according to Abbe’s law, it is dAbbe = λ/2NA, where λ is the used laser wavelength and NA is the numerical aperture of the objective lens. For visible radiation and high NA objectives, dAbbe is ~0.300 μm in the transverse plane, while in the axial direction it is ~1.2 μm. It is clear that, for extended samples, it is possible to acquire the Raman spectra on a two- or three-dimensional array of points, with a step comparable with the spatial resolution (raster scan). The collected spectra can then be analyzed in terms of the intensity of a selected Raman band corresponding to a specific bio-chemical component of the sample. The result is a spatial image of the abundance of that specific component (Raman imaging). Nowadays, Raman imaging is becoming a tool for a patchwork of interdisciplinary research, involving physicists, chemists, biologists, and molecular biologists and clinicians. Alongside the many advantages of RS, however, it should be emphasized that the cross section of Raman scattering is quite low, typically ~10−25−10−30 cm2, depending on whether the process is resonant or non-resonant. In a typical Raman measurement using a diffraction-limited laser beam, the typical detection volume is of the order of few fL and, for concentrations of ~1 μM, the total number of scattering molecules is ~106. Even after optimizing the power of the Raman probe and the integration time, the obtained signal approaches a signal-to-noise ratio close to 1, i.e., clearly not enough for many applications in which the analytes are present at sub μM concentrations, or for very thin samples. As will be discussed in the next paragraph, this drawback has been overcome thanks to the discovery of the SERS (surface-enhanced Raman scattering) technique which pushes the limit of detection even to the single molecule level [30][23].3. Surface- and Tip-Enhanced Raman Scattering
The discovery of SERS dates backs to 1974, when Fleishmann and co-workers [17][24] accidentally observed an anomalous enhancement of the Raman signal when pyridine molecules were deposited over a roughened silver surface. A few years later, two independent papers [31,32][25][26] provided a physical interpretation of the phenomenon based on localized surface plasmon resonances (LSPRs) induced by the laser radiation shining on metal nanostructured surfaces. Since this initial discovery, there has been an impressive growth in the number of publications that have confirmed this phenomenon, and the SERS technique is today a powerful spectroscopic tool with high sensitivity and chemical selectivity [33,34,35][27][28][29]. Research activity in the field of plasmon-enhanced spectroscopies and their numerous applications is nowadays rooted in many laboratories scattered around the world. An indicator of the great interest in this research topic is the huge number of publications: by searching various databases using the keyword SERS more than 3 × 104 papers are found. Even in Italy, there is very lively activity on this topic, but the detailed description of these studies is beyond the scope of this text. For more information on the trend of Italian research, one can visit the website www.plasmonica.it (accessed on 14 September 2023). Among the countless applications, we note the most significant ones: environmental science [43][30], cancer detection [44][31], free immunoassay platform [45][32], food safety [46][33], and, very recently, coronavirus detection [47][34]. Coming to the basic physical principles, LSPR originates from the interaction between laser radiation and free electrons, which causes coherent and collective electron oscillations at the interface of two media having the real part of dielectric functions of the opposite sign. This condition is usually realized at the interface between noble metals (mainly gold and silver) and dielectrics. Noble metals exhibit resonance wavelength located typically in the visible and near-infrared regions. Although metals are the most used plasmonic materials, significant plasmonic responses are also observed in heavily doped semiconductors and 2D materials. The plasmon frequency is certainly the most representative parameter of plasmonic effect: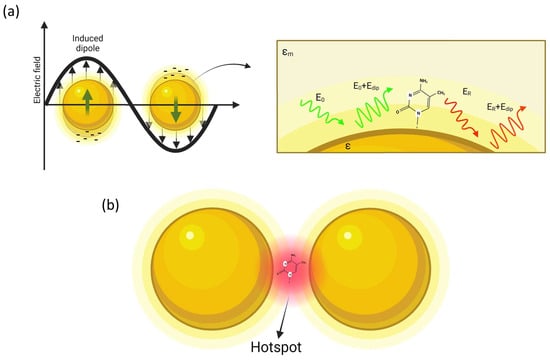
Figure 1. Simple description of SERS: (a) a metal nanoparticle illuminated by light causes the oscillation of the free electrons of the surface, so that a molecule placed at nanometric distance from the NP surface is affected by an amplified optical field; (b) amplification is further increased in the gap region in a dimer structure (hot-spot).
References
- Keiser, G. Biophotonics Concept and Applications, 1st ed.; Springer: Singapore, 2016.
- Prasad, P.N. Introduction to Biophotonics; Wiley: Hoboken, NJ, USA, 2003.
- Tsia, K. Understanding Biophotonics: Fundamentals, Advances, and Applications, 1st ed.; CRC Press: Boca Raton, FL, USA, 2015.
- Pavesi, L.; Fauchet, P. Biophotonics; Springer-Verlag: Berlin/Heidelberg, Germany, 2008.
- Meglinski, I. Biophotonics for Medical Applications, 1st ed.; Woodhead Publishing: Sawston, UK, 2015.
- Jensen, E.C. Types of imaging, part 2: An overview of fluorescence microscopy. Anat. Rec. 2012, 295, 1621–1627.
- Smith, C.L. Basic Confocal Microscopy. Curr. Protoc. Neurosci. 2011, 56, 2.
- Wäldchen, S.; Lehmann, J.; Klein, T.; van de Linde, S.; Sauer, M. Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 2015, 5, 15348.
- Raman, C.V. A change of wavelength in light scattering. Nature 1928, 121, 619.
- Krafft, C.; Popp, J. The many facets of Raman spectroscopy for biomedical analysis. Anal. Bioanal. Chem. 2015, 407, 699–717.
- Kumar, C.S.S.R. Raman Spectroscopy for Nanomaterials Characterization, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2012.
- Gregoriou, V.G.; Braiman, M.S. Vibrational Spectroscopy of Biological and Polymeric Materials, 1st ed.; CRC Press: Boca Raton, FL, USA, 2006.
- Siebert, F.; Hildebrandt, P. Vibrational Spectroscopy in Life Science; Wiley: Hoboken, NJ, USA, 2008.
- Lasch, P.; Kneipp, J. Biomedical Vibrational Spectroscopy; Wiley-Interscience: Hoboken, NJ, USA, 2008.
- Rusciano, G.; Capriglione, P.; Pesce, G.; Carnovale, V.; Sasso, A. Raman spectroscopy as a new tool for early detection of bacteria in patients with cystic fibrosis. Laser Phys. Lett. 2013, 10, 075603.
- Qi, Z.; Akhmetzhanov, T.; Pavlova, A.; Smirnov, E. Reusable SERS Substrates Based on Gold Nanoparticles for Peptide Detection. Sensors 2023, 23, 6352.
- Gremlichl, H.-U.; Bing, Y. Infrared and Raman Spectroscopy of Biological Materials, 1st ed.; CRC Press: Boca Raton, FL, USA, 2001.
- Butler, H.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687.
- Rusciano, G.; De Luca, A.C.; Pesce, G.; Sasso, A. Phase-sensitive detection in Raman Tweezers. Appl. Phys. Lett. 2006, 89, 261116.
- Rusciano, G.; De Luca, A.C.; Pesce, G.; Sasso, A. Enhancing Raman tweezers by phase-sensitive detection. Anal. Chem. 2007, 79, 3708–3715.
- De Luca, A.C.; Rusciano, G.; Ciancia, R.; Martinelli, V.; Pesce, G.; Rotoli, B.; Sasso, A. Resonance Raman spectroscopy and mechanics of single red blood cell manipulated by optical tweezers. Haematologica 2007, 92, 174.
- De Luca, A.C.; Rusciano, G.; Ciancia, R.; Martinelli, V.; Pesce, G.; Rotoli, B.; Sasso, A. Spectroscopical and mechanical characterization of normal and thalassemic red blood cells by Raman tweezers. Opt. Exp. 2008, 16, 7943–7957.
- Kneipp, K.; Kneipp, H.; Kartha, V.B.; Manoharan, R.; Deinum, G.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Detection and identification of a single DNA base molecule using surface-enhanced Raman scattering (SERS). Phys. Rev. E 1998, 57, R6281–R6284.
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166.
- Jeanmaire, D.L.; Van Duyne, R.P. Surface Raman spectroelectrochemistry; Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. 1977, 84, 1–20.
- Albrecht, M.G.; Creighton, J.A. Anomalously intense Raman spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217.
- Lin, D.-Y.; Yu, C.-Y.; Ku, C.-A.; Chung, C.-K. Design, fabrication, and applications of SERS substrates for food safety detection: Review. Micromachines 2023, 14, 1343.
- Beeram, R.; Vepa, K.R.; Soma, V.R. Recent trends in SERS-based plasmonic sensors for disease diagnostics, biomolecules detection, and machine learning techniques. Biosensors 2023, 13, 328.
- Jebakumari, K.A.E.; Murugasenapathi, N.K.; Palanisamy, T. Engineered two-dimensional nanostructures as SERS substrates for biomolecule sensing: A review. Biosensors 2023, 13, 102.
- Ong, T.T.X.; Blanch, E.W.; Jones, O.A.H. Surface enhanced Raman spectroscopy in environmental analysis, monitoring and assessment. Sci Total Environ. 2020, 720, 137601.
- Qian, X.; Peng, X.H.; Ansari, D.O.; Yin-Goen, Q.; Chen, G.Z.; Shin, D.M.; Yang, L.; Young, A.N.; Wang, M.D.; Nie, S. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008, 26, 83–90.
- Rusciano, G.; Capaccio, A.; Sasso, A.; Capo, A.; Murillo Almuzara, C.; Staiano, M.; D’Auria, S.; Varriale, A. A SERS-based biosensor for the detection of biological macro-molecules: The case of the lipopolysaccharide endotoxin molecules. Int. J. Mol. Sci. 2023, 24, 12099.
- Xu, M.L.; Gao, Y.; Han, X.X.; Zhao, B. Innovative application of SERS in food quality and safety: A brief review of recent trends. Foods 2022, 11, 2097.
- Peng, Y.; Lin, C.; Li, Y.; Gao, Y.; Wang, J.; He, J.; Huang, Z.; Liu, J.; Luo, X.; Yang, Y. Identifying infectiousness of SARS-CoV-2 by ultra-sensitive SnS2 SERS biosensors with capillary effect. Matter 2022, 5, 694–709.
- Le Ru, E.C.; Etchegoin, P.G. Principles of Surface-Enhanced Raman Spectroscopy and related Plasmonic Effects, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2009.
- Kneipp, K.; Moskovits, M.; Kneipp, H. Surface-Enhanced Raman Scattering: Physics and Applications, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2006.
- Anderson, D.J.; Moskovits, M. A SERS-active system based on silver nanoparticles tethered to a deposited silver film. J. Phys. Chem. B 2006, 110, 13722–13727.
- Lin, C.; Liang, S.; Peng, Y.; Long, L.; Li, Y.; Huang, Z.; Long, N.V.; Luo, X.; Liu, J.; Li, Z.; et al. Visualized SERS imaging of single molecule by Ag/Black phosphorus nanosheets. Nano-Micro Lett. 2022, 14, 75.
- Cong, S.; Liu, X.; Jiang, Y.; Zhang, W.; Zhao, Z. Surface enhanced Raman scattering revealed by interfacial charge-transfer transitions. Innovation 2020, 1, 100051.
- Le Ru, E.C.; Etchegoin, P.G.; Meyer, M. Enhancement factor distribution around a single surface-enhanced Raman scattering hot spot and its relation to single molecule detection. J. Chem. Phys. 2006, 125, 204701.
- Stöckle, R.M.; Suh, Y.D.; Deckert, V.; Zenobi, R. Nanoscale chemical analysis by tip-enhanced Raman spectroscopy. Chem. Phys. Lett. 2000, 318, 131–136.
- Anderson, M.S. Locally Enhanced Raman Spectroscopy with an Atomic Force Microscope. Appl. Phys. Lett. 2000, 76, 3130–3132.
- Hayazawa, N.; Inouye, Y.; Sekkat, Z.; Kawata, S. Metallized tip amplification of near-field Raman scattering. Opt. Commun. 2000, 183, 333–336.
- Pettinger, B.; Ren, B.; Picardi, G.; Schuster, R.; Ertl, G. Nanoscale probing of adsorbed species by tip-enhanced Raman spectroscopy. Phys. Rev. Lett. 2004, 92, 096101.
- Itoh, T.; Procházka, M.; Dong, Z.C.; Ji, W.; Yamamoto, Y.S.; Zhang, Y.; Ozaki, Y. Toward a new era of SERS and TERS at the nanometer scale: From fundamentals to innovative applications. Chem. Rev. 2023, 123, 1552–1634.
- Bharadwaj, P.; Deutsch, B.; Novotny, L. Optical Antennas. Adv. Opt. Photonics 2009, 1, 438–483.
- Fan, Y.; Jin, D.; Wu, X.; Fang, H.; Yuan, X. Facilitating hotspot alignment in tip-enhanced Raman spectroscopy via the silver photoluminescence of the probe. Sensors 2020, 20, 6687.
- Taguchi, A.; Yu, J.; Verma, P.; Kawata, S. Optical antennas with multiple plasmonic nanoparticles for tip-enhanced Raman microscopy. Nanoscale 2015, 7, 17424–17433.
- Novotny, L.; Van Hulst, N. Antennas for light. Nat. Photonics 2011, 5, 83–90.
- Rusciano, G.; Zito, G.; Isticato, R.; Sirec, T.; Ricca, E.; Bailo, E.; Sasso, A. Nanoscale chemical imaging of bacillus subtilis spores by combining Tip-Enhanced Raman Scattering and advanced statistical tools. ACS Nano 2014, 8, 12300–12309.
- Huang, T.X.; Cong, X.; Wu, S.S.; Lin, K.Q.; Yao, X.; He, Y.H.; Wu, J.B.; Bao, Y.F.; Huang, S.C.; Wang, X.; et al. Probing the edge-related properties of atomically thin MoS2 at nanoscale. Nat. Commun. 2019, 10, 5544.
- Schultz, J.F.; Mahapatra, S.; Li, L.; Jiang, N. The expanding frontiers of tip-enhanced Raman spectroscopy. Appl. Spectrosc. 2020, 74, 313–1340.
- Novotny, L.; Stranick, S.J. Near-field optical microscopy and spectroscopy with pointed probes. Annu. Rev. Phys. Chem. 2006, 57, 303–331.
- Schmid, T.; Opilik, L.; Blum, C.; Zenobi, R. Nanoscale chemical imaging using tip-enhanced Raman spectroscopy: A critical review. Angew. Chem. Int. Ed. 2013, 52, 5940–5954.
- Stadler, J.; Schmid, T.; Zenobi, R. Developments in and practical guidelines for tip-enhanced Raman spectroscopy. Nanoscale 2012, 4, 1856–1870.
- Deckert, V.; Deckert-Gaudig, T.; Diegel, M.; Götz, I.; Langelüddecke, L.; Schneidewind, H.; Sharma, G.; Singh, P.; Zeisberger, M.; Zhang, Z.; et al. Spatial resolution in Raman spectroscopy. Faraday Discuss. 2015, 177, 9–20.
- Zhang, R.; Zhang, Y.; Dong, Z.C.; Jiang, S.; Zhang, C.; Chen, L.G.; Zhang, L.; Liao, Y.; Aizpurua, J.; Luo, Y.; et al. Chemical mapping of a single molecule by plasmon-enhanced Raman scattering. Nature 2013, 498, 82–86.
- Verma, P. Tip-enhanced Raman spectroscopy: Technique and recent advances. Chem. Rev. 2017, 117, 6447–6466.
- Cao, Y.; Sun, M. Tip-enhanced Raman spectroscopy. Rev. Phys. 2022, 8, 100067.
- Shi, X.; Coca-López, N.; Janik, J.; Hartschuh, A. Advances in Tip-enhanced near-field Raman microscopy using nanoantennas. Chem. Rev. 2017, 117, 4945–4960.
- Langeluddecke, L.; Singh, P.; Deckert, V. Exploring the nanoscale: Fifteen years of tip enhanced Raman spectroscopy applied spectroscopy. Appl. Spectrosc. 2015, 69, 1357–1371.
- Kumar, N.; Mignuzzi, S.; Su, W.; Roy, D. Tip-enhanced Raman spectroscopy: Principles and applications. EPJ Tech. Instrum. 2015, 2, 9.
- Pienpinijtham, P.; Kitahama, Y.; Ozaki, Y. Progress of tip-enhanced Raman scattering for the last two decades and its challenges in very recent years. Nanoscale 2022, 14, 5265–5288.
- Kausar, A.S.M.Z.; Reza, A.W.; Latef, T.A.; Ullah, M.H.; Karim, M.E. Optical nano antennas: State of the art, scope and challenges as a biosensor along with human exposure to nano-toxicology. Sensors 2015, 15, 8787–8831.
- Bartolomeo, G.L.; Goubert, G.; Zenobi, R. Tip recycling for atomic force microscopy-based tip-enhanced Raman spectroscopy. Appl Spectrosc. 2020, 74, 1358–1364.
More
