Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Islam EL JADDAOUI.
Libraries contain a large amount of organic material, frequently stored with inadequate climate control; thus, mold growth represents a considerable threat to library buildings and their contents.
- library
- filamentous fungi
- mold
- mould
- mold isolation
1. Introduction
When we speak of libraries, we mean both the books, documents, and other materials that constitute a collection as well as the buildings that house these materials. Perhaps the most famous quote about libraries comes from Argentinian author Jorge Luis Borges (1899–1986), who said, “I have always imagined that Paradise will be a kind of library.” (“Siempre imaginé que el Paraíso sería algún tipo de biblioteca.”) [1].
Throughout history, the biggest threat to libraries has come from fires [2]. These fires have been a side effect of war, set deliberately by conquerors, or as collateral damage during bombing. Sometimes, religious fanatics who have feared the secular knowledge stored within libraries create purposeful conflagrations; in other cases, fires occur through accidents or poor management of the physical plants in which library collections are housed [3,4,5][3][4][5].
Fungal damage to library materials is often first suspected because of the musty odors that are caused by volatile organic compounds (VOCs) produced during the biodeterioration process (Figure 1) [6]. Fungi also increase the rate of paper deterioration through foxing. The color of the fox’s spots can originate from two sources: a rust-red alkali-soluble material and an alkali-insoluble straw-colored stain in the paper fibers. When contaminated materials are new, there is no noticeable discoloration, but the fungal structures or products form a suitable chemical environment that, over time, causes discoloration [7]. More dramatically, lush and easily observed mold growth can be a serious aftereffect of fire events. When fires are discovered early enough for fire departments to succeed in putting them out, in some cases, the ensuing water and microbial damage constitutes a major part of the loss of books [8]. Most commonly, however, water in libraries is due to intrusion from floods, structural leaks, or condensation when moist warm air encounters a cold surface. In humid climates where air conditioning is widespread, condensation can be a major problem despite good building maintenance. Outdoor fungal aerosols often dominate indoor fungal populations, even with complex ventilation systems. When sufficient water is present, dust that accumulates in carpeting and ventilation duct surfaces can become important inoculum sources [9,10][9][10].
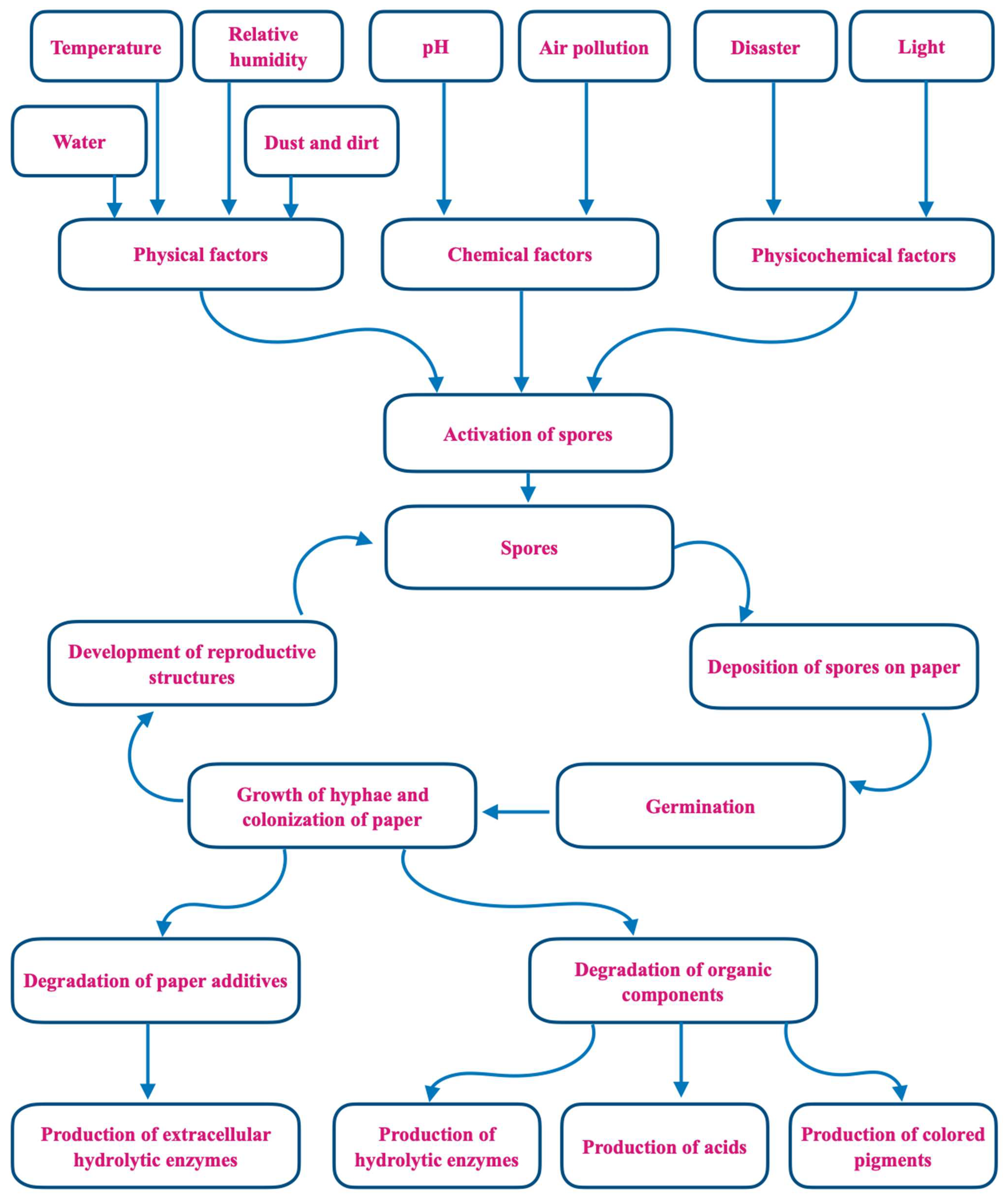
Figure 1. Mechanisms of fungal biodeterioration of paper.
Climate change and global warming have increased the threat of extreme storms, flooding, and other weather events that lead to water incursion in buildings. Although libraries constitute only one type of built environment, their contents make them particularly susceptible to microbial biodeterioration. In addition to paper, volume bindings made of cardboard, fabric, leather, and glue are all good substrates for mold growth [11,12][11][12].
2. Methods for Sampling and Identification of Fungi
2.1. Sampling Methods
The methods used for sampling indoor fungi have been developed and most widely used in the food and pharmaceutical industries or hospital settings [21][13] and then adapted for use in other indoor environments. The manual published by the American Industrial Hygiene Association (AIHA) titled “Recognition, Evaluation and Control of Indoor Mold” [22][14] is one of the best single sources for guidance on the evaluation and analysis of indoor molds. Other valuable resources include Chin and Heinsohn [23][15], Flannigan et al. [24][16], Verdier et al. [25][17], and Martin-Sanchez et al. [26][18]. Many different methods developed for the study of indoor environments have been used to assess mold damage in libraries (Figure 2). These include culturing, mechanical air samplers, spore counting, direct microscopy, and DNA and chemical analyses [22,24,25,26][14][16][17][18]. Traditional sampling is conducted either from the air, dust, books, shelving, and other surfaces or—when evident—directly from fungal growth [27][19]. A diagrammatic overview of different traditional sampling techniques is given in Figure 3.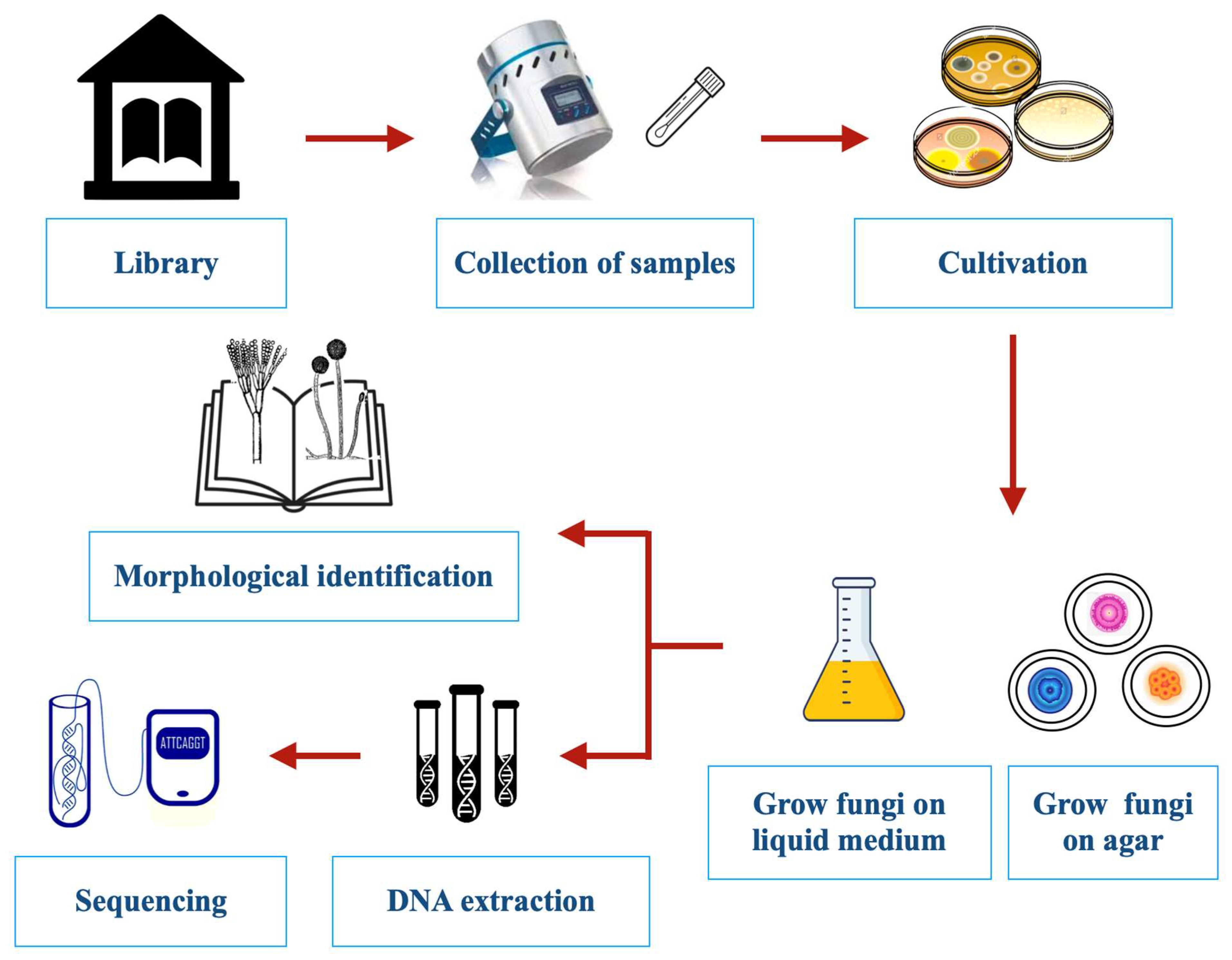
Figure 2. Experimental protocols for the detection, isolation, and identification of fungi colonizing libraries.
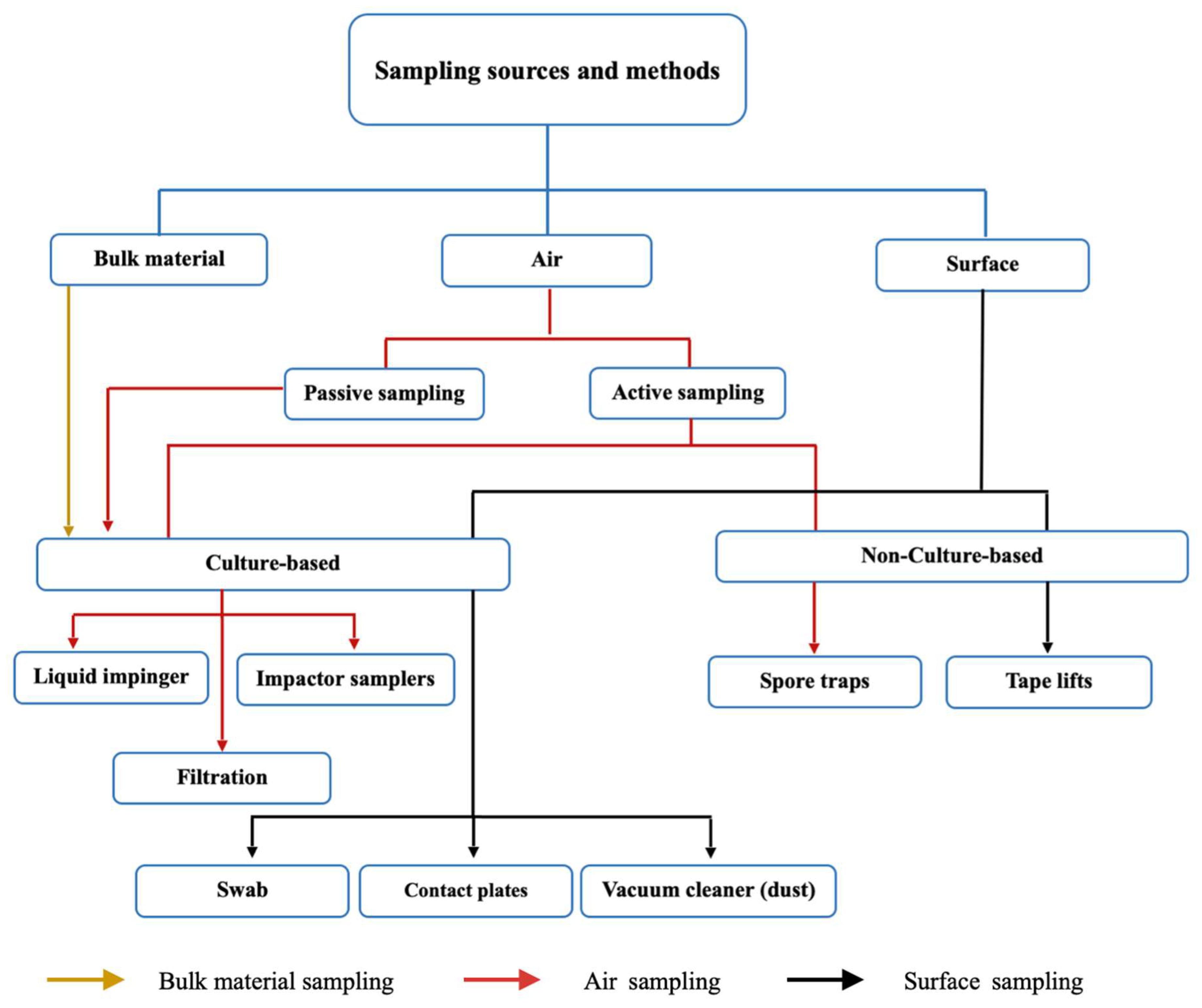
Figure 3. Conventional sampling strategies for isolating, enumerating, and identifying fungi from indoor environments.
2.1.1. Air Sampling
Choosing the proper air sampling strategy depends on several factors, including indoor environmental conditions, the purpose of the research and analysis to be performed, and the concentration of microorganisms in the air. Most active air sampling equipment consists of a minimum of a sampling pump with an airflow inlet and includes the use of a collection medium. Air sampling also relies on certain parameters such as constant and appropriate flow rates and stable ambient conditions [28][20]. With culture-based air sampling, the concentrated particulates are then impacted onto an agar growth medium. This sampling method is widely used for determining the presence and concentration of culturable fungal spores and fragments. Quantifiable data from viable air sampling methods are reported as colony-forming units (CFUs) per unit volume of air sampled. Specialized devices have been developed for the collection of fungal propagules from the air. In a standard impactor sampler, the air is accelerated through a perforated plate or a narrow slit after being drawn into a sampling head with a pump or fan. This creates laminar airflow over the collecting surface, which is frequently a standard agar plate or contact plate filled with suitable agar medium. The size of the pores in sieve samplers and the width of the slit in slit samplers affect the air’s velocity. When air hits the surface of the collection, it produces a tangential variation in direction, and any suspended particles are carried away by inertia, impacting the surface of the collection [29][21]. A known volume of air is passed through the device at a defined flow rate for a predetermined amount of time. The prototype for this form of sampling was developed by Andersen in 1958 (Ariel A. Andersen, Provo, UT, USA), and many subsequent refinements and improvements have been made since then [22][14]. A wide range of devices have been developed using the impaction principle. The Andersen sampler, a multi-stage cascade sieve device that uses perforated plates with progressively smaller holes at each level, is one of the most commonly used. This instrument allows particles to be separated based on their size [28][20]. Impingement-based methods work almost the same as impact-based techniques, except that the microorganisms are collected in a liquid medium. Typically, air is drawn through a narrow inlet tube over the collection medium, where the flow rate of sampled air depends on the diameter of the inlet nozzle. The suspended particles are projected onto the collecting liquid as soon as the air hits the liquid. At the end of sampling, aliquots are grown on appropriate growth media to enumerate viable microorganisms [30][22]. Impinger sampling is particularly useful for sampling heavily contaminated air because liquid samples can be diluted to the appropriate level for subsequent growth culture analysis [31][23]. Liquid impingers (The United States Bureau of Mines Experiment Station, Pittsburgh, PA, USA) are more practical for post-analysis, as biological agents present in liquid media would not be dehydrated. However, the minimum particle cutoff size is 300 nm for commercial impingers, and they have shown very low collection efficiency for particles smaller than 100 nm [32][24]. Filtration-based samplers are relatively less complicated and less expensive for sampling bioaerosols. In the filtration method, airborne microorganisms are collected by passing air through porous membrane filters made of fiberglass, polycarbonate, polyvinyl chloride, or cellulose acetate and incubated by transfer onto the surfaces of growth agar medium or in gelatin [31][23]. Spore traps are used for non-culture-based analysis. Like culture-based fungal air sampling, these devices rely on the impaction of suspended airborne particles moving at a relatively high velocity. Rather than a growth medium, glass slides with a sticky transparent material or a sticky transparent membrane are used to collect the spores, which can then be counted and examined under an optical microscope [22][14]. The use of spore-trapping samplers to collect fungal spores, followed by direct microscopy to identify and enumerate the spores, facilitates an estimate of the total number of particulates present in a sample. Sometimes, microscopic stains such as basic fuchsin or lactophenol cotton blue are used to discriminate spores from simple debris [33][25]. The Burkard spore trap (Rothamsted Experimental Station, Harpenden, UK) is widely used, and samples collected with this instrument are analyzed using microscopy for spore identification, usually to the genus level [34][26]. However, because only a small number of fungal spore types have distinctive morphologies, this approach only gives a rough idea of the kinds of fungi that are present. Aspergillus and Penicillium are often the predominant fungi detected from indoor air, including that of libraries. Under the light of a microscope, the spores of both genera are seen as small undifferentiated spheres. Thus, non-culture-based microscopic methods do not allow for the differentiation between genera of Aspergillus and Penicillium or within species of either genus. In other cases, fungi can be identified, at least to genera, by their spore shape. Cladosporium, Alternaria, and Stachybotrys are examples of genera with distinctive spore morphology. For taxa with morphologically uninformative spore morphology, culture-based methods offer a practical and convenient way to observe enough characters to identify species [35][27].2.1.2. Surface Sampling
For surface sampling, swab sampling is the most commonly used. In this method, a sterile swab is rubbed over the surface of interest using a twisting motion. The swab is then subjected to microbial examination, usually by shaking it in a diluent and examining the resulting microbial suspension using pour plate cultures. Swabs are convenient for use on irregular surfaces, where contact plates cannot be used [36][28]. A contact plate or Rodac (VWR International, Radnor, PA, USA) is an agar poured into a contact plate usually used for surface sample testing. The process begins by removing the lid and gently rolling the agar surface across the sample area, transferring any microorganisms present on the surface to the agar. After obtaining the sample, the cover is replaced, and the surface is cleaned with a wipe and isopropyl alcohol to remove any agar residue remaining from the contact plate. The plates are then incubated at temperatures suitable for the growth of a variety of microorganisms [36][28]. Adhesive tape (IECL, Brisbane, Australia) sampling is a non-destructive technique that allows the determination of which fungal particles have accumulated on a specific surface. Although tape sampling and direct microscopic examination cannot determine which spores are viable, they can identify some species regardless of their capacity to develop on a specific culture media. To aid proper observation of the sample, the piece of tape must be placed in the center of the microscope slide [22][14]. This is a simple and inexpensive technique that does not require any specific equipment and shows the existing relationships between the surface and the colonizing microorganisms, the diffusion, and the correspondence with a particular alteration of the surface.2.1.3. Factors Affecting Sampling
The season of the year, building characteristics such as construction materials, air conditioning system, ventilation, and light source as well as other environmental parameters, mainly indoor temperature and relative humidity, can affect the data collected [37][29]. The main fungi recovered from a particular environment will partially reflect the analytical method used to detect them [38][30]. Each sampling strategy and analytical method inevitably involves creating a bias. Fungal counts in floor dust are less affected by short-term variability than airborne concentrations and thus have the advantage of providing an estimate of mold quantities over a given period. Furthermore, certain mechanical samplers are better for certain purposes. The Andersen six-stage sampler is preferred for viable counts, and the Burkard 24 h sampler is preferred for total counts [39][31]. When studying viable counts, different types of media recover different types of organisms. For example, when media with low water activity are used, they will select for xerophilic fungi [40][32]. Moreover, all sampling methods based on cultivation will find only viable fungal spores. In the 21st century, PCR has emerged as an effective way of detecting slow-growing species as well as non-viable propagules. A gradual shift toward DNA-based methods has taken place in recent decades [26,41][18][33].2.1.4. Culture-Based and Non-Culture-Based Sampling
The major disadvantage of culture-based sampling over non-culture-based sampling is that it tests only for viable propagules; moreover, not all viable fungal propagules necessarily germinate on the selected growth medium. Certain fungi grow quickly, while others grow slowly. It is possible that one species of quickly growing fungus will dominate and overgrow a nutrient agar plate before other slow-growing types can develop. This means that interpreting data from such samples can be difficult. More importantly, culturable organisms comprise a small fraction of the total number of fungi found in each sample [35,42][27][34]. Despite these limitations, culture-based fungal air sampling is still a widespread and useful investigative tool [22][14]. Moreover, by combining techniques, some of the limits of each methodology are addressed, and many researchers use at least two approaches for any given sample [43][35]. The paper by Nazaroff [44][36] provides useful insights into the dynamic processes that govern bioaerosol behavior and possible impacts on human health.2.2. Fungal Identification Techniques
2.2.1. Microscopic Identification
There is a shortage of skilled mycologists who can accurately identify species using traditional microscopic and cultural approaches. Environmental microbiologists have developed alternatives to counting and culturing for the identification of fungi. These include the detection of ergosterol content [45][37], beta-glucans [46][38], nucleic acids [47][39], and VOCs [48][40] as well as immunoassays [49][41]. In the majority of cases, molds are identified based on colony and microscopic characteristics. In-depth microscopic analysis of a fungal specimen yields a clear picture and useful details on the fungal structure. Thus, experts can typically identify molds with certainty based on their microscopic structures and morphological characteristics, such as type, size, shape, and arrangement of their spores, as well as their hyphae’s size, color, and septation. However, unlike bacterial and yeast identification, species-level mold identification has not been subject to the same formal standards. A combination of macroscopic and microscopic examination can be used to identify molds. The first step in the identification of fungal specimens is the use of a definite fungal stain. The selection of the staining method is primarily based on the sample under study [50][42].2.2.2. Molecular Identification
The single best substitute for counting viable and non-viable colonies is PCR because this technique offers several advantages over the more time-consuming traditional monitoring strategies. It can usually identify fungi to the species level while also decreasing the time required for the more time-consuming and less sensitive culture and counting methods. In addition, quantitative PCR can reduce the variability associated with culture-based methods [51][43]. Mycologists have developed a system of “DNA barcoding” that facilitates the rapid identification of fungi [52][44]. Using such culture-independent nucleic acid analysis, Amend et al. [53][45] found a high diversity of fungi from global indoor environments with Alternaria, Cladosporium, and Epicoccum as well-represented as the Aspergillus and Penicillium species that tend to dominate culture-based surveys. When several different indoor buildings in Helsinki were studied with this approach, 585 fungi were identified, a much larger number than is usually reported using traditional cultural and microscopic analysis indicating that there is more diversity in indoor fungal populations than has been previously identified [54][46]. Two primary methods can be used in real-time PCR and traditional PCR to identify the precise target sequences required for the detection and identification of fungal strains: (1) using universal primers to amplify and sequence conserved genes shared by all fungus and (2) using non-specific random primers to amplify unknown genomic regions. The highly stable, varied, and conserved sequences of the nuclear-encoded ribosomal RNA genes (rRNA), as well as their ability to be amplified and sequenced using universal primers, make them desirable targets. They also occur in multiple copies arranged in tandem repeats, with each repeat made up of the 18S small subunit (SSU), 5.8S large subunit, and 28S large subunit (LSU) genes separated by internal transcribed spacer (ITS) regions (ITS1 and ITS2). Another part of rDNA is the spacer between the LSU and SSU genes, called the nontranscribed spacer (NTS) or intergenic spacer (IGS). The rDNA-conserved regions enable the detection of similar ribosomal genes from various species, genera, families, or even kingdoms using probes or primers from a single species. However, there are numerous sequence variations among the identical DNA fragments found in different organisms that might be used to identify them [55][47]. The key fungal DNA barcode for species identification is the ITS region. The primary benefit of the ITS region is the simplicity of PCR amplification from small samples using universal primers. Furthermore, the ITS region has generally high PCR amplification efficacy rates. Another significant advantage of the ITS is the abundance of high-quality reference sequences that have been deposited in numerous online databases [56][48]. The majority of sequence variation in rDNA exists within the IGS regions. IGS regions are more challenging to amplify and sequence than ITS regions, but they can be helpful when there are not enough variations among ITS regions. Ascomycete mating-type genes, the elictin, and the laccase gene are other genes that are used as detection targets. Specific target sequences can also be identified by amplifying random regions of the fungal genome with PCR-based strategies such as arbitrarily primed PCR (AP-PCR) and randomly amplified polymorphic DNA (RAPD). Amplified PCR fragments are separated with gel electrophoresis to identify unique PCR bands that can be purified, cloned, and sequenced. Compared with the amplification of conserved genes, this method requires more time and expertise because it necessitates the study of several isolates from both the target species and its related species [55][47]. While we recognize that until recently, these methods were rarely used in the context of libraries [57][49], we recommend that in the future, librarians who seek to learn about mold contaminants in their collections should use molecular methods. In the future, DNA-based approaches will become the “gold standard” and will allow more precise and accurate identification of fungal taxa. High-throughput DNA sequencing can facilitate a better characterization of microbial communities. Sequence-based approaches can be used to extend or replace culture- or microscopy-based techniques for identifying indoor fungi and can often give direct identification of species [58][50]. Ideally, a combination of molecular and morphological techniques to provide accurate species identifications will enable researchers to better evaluate the potential for toxin and allergen production [59][51]. Obviously, there is a great need for standardizing the methods used for assessing microbial contamination in the built environment. Because so many different approaches are used by different laboratories in different countries, it is difficult to compare findings from different laboratories.2.2.3. Fourier Transform Infrared (FTIR) Spectroscopy Identification
Conventional phenotypic methods for fungal identification are time-consuming, and most alternative methods are still expensive and call for specialized laboratory knowledge. It has been demonstrated during the past two decades that FTIR spectroscopy is a useful approach for identifying microorganisms. FTIR is a non-destructive, robust [60][52], and vibrational spectroscopic technique based on measuring the fundamental vibrational modes of a molecular bond. In this method, a sample interacts with a polychromatic infrared source, and the molecules therein can either absorb or reflect the light, stimulating vibrational motions [61][53]. The foundation of FTIR is the capture of each microorganism’s unique spectral signature after cultivation under predetermined conditions. This spectral signature primarily reflects the biomass’s composition in terms of proteins, lipids, nucleic acids, and carbohydrates [62][54]. The research conducted by Lecellier et al. [61][53] validates the efficacy of high throughput FTIR spectroscopy for the identification of fungi utilizing a library of molds with industrial significance [61][53]. Foxing spots were also identified using FTIR spectroscopy on paper from nine printed books that ranged in age from the early 19th to the mid-20th century [60][52].2.2.4. VOCs Identification
As we already mentioned, mycologists also have used VOCs to detect molds. VOCs are carbon-containing compounds of low molecular mass that easily evaporate at normal temperature and pressure. Most microbial VOCs have distinct smells. More than 300 VOCs from microscopic and macroscopic fungi have been reported, and it is usual for dozens, if not hundreds, of fungal species to release the same chemical type of VOC [63][55]. VOCs are metabolic byproducts of fungi that can be detected before apparent symptoms of microbial growth [64][56]. As a result, this fingerprint can act as early warning signs of potential biocontamination issues in library materials. The analysis of VOCs produced by molds developed in libraries has been addressed by several studies [65,66,67][57][58][59].2.2.5. Scanning Electron Microscopy (SEM) Identification
In addition to VOCs, SEM and energy-dispersive X-ray spectroscopy (EDS) are also used as fungi detection techniques. SEM/EDS comprise what has long been considered the advanced surface examination tool for materials scientists. SEM is the imaging portion of the technique. A scanning electron microscope, in contrast with a standard optical microscope, converts electron interactions into an optical signal using electrons. Despite the benefits of optical microscopy for some applications, there may be resolution restrictions caused by limited focus depth and light wavelength [68][60]. For the investigation of fungi, the scanning electron microscope is perfectly suited for the observation of intact spore structures over a wide range of magnifications, duplicating and supplementing data obtained using light microscopy [69][61].2.2.6. Biochemical Identification
In certain cases, fungal identification may require biochemical tests that distinguish genera among families and species among genera. For the identification of various molds, numerous common biochemical assays are available, such as urease production and proteolysis. When fungi grow in selective solid or liquid media, they ferment carbohydrates and produce alcohols, acids, gases, and enzymatic and metabolic products in patterns characteristic of their genus and/or species. These fermentation byproducts can differentiate between taxa [50][42].3. Mold Prevention in Libraries
The protection of library materials can be optimized by promoting dry, stable environmental conditions, which, in most climates, means temperature-controlled heating and air conditioning systems. Suitable environmental parameters include setting the temperature as low as possible to reduce temperature-induced deterioration and controlling relative humidity to avert damage to the collection [70,71][62][63]. Ideally, each library should have a temperature and humidity monitoring system because if one of these parameters in the building falls outside acceptable limits, mold will grow on library materials [72][64]. Norms for indoor relative humidity and temperature broadly depend on local climatic conditions, which is why these norms cannot always be considered standards. It is crucial that librarians are aware of the different prerequisites. It is also important to know that a well-installed and maintained air conditioning system is not enough, because the system itself cannot compensate for design and construction defects. The structure of a building must create an airtight environment that is then complemented with an air conditioning system. Implementation of a Heating, ventilation, and air conditioning (HVAC) system sometimes has negative effects, especially with relative humidity [73][65]. Happily, monitoring library temperature and humidity can be accomplished with a very reasonable investment in a digital thermohygrometer [72][64]. For libraries in many tropical countries, these climatic control systems are too expensive, leading to a periodic appearance of mold growth [74][66]. Moreover, in subtropical and tropical climates, air conditioning is usually installed more for human comfort than for the protection of library collections. When these libraries are closed, the systems are often turned off, resulting in wide temperature fluctuations that can lead to water condensation and subsequent mold growth. Several experts have emphasized that maintaining the indoor environment at certain temperatures and relative humidity levels is effective in reducing the propagation of fungi and other microorganisms [37,75,76][29][67][68]. Ventilation lowers levels of existing mold spores in the air while keeping the environment dry and cool. HVAC systems with good system design can provide environmental control over large regions of a building. Proper equipment maintenance also decreases the likelihood of issues caused by system failures [77][69]. Control of indoor mold is a multidisciplinary problem. Many environmental variables, including occupancy, building structure, maintenance, ventilation, and climate, affect the mycology of a given library. Mycologists should become more aware of the pitfalls associated with the traditional methods of screening for fungi and the challenges that accompany attempts to compare studies across climates, library characteristics, sampling methods, and the like. Proper use and monitoring of HVAC systems minimize mold contamination problems, but the single most important preventative measure is keeping water out by ensuring that roofs do not leak and that indoor plumbing remains in good repair. Librarians should become more aware of the guidelines issued by the “American Society of Heating, Refrigerating and Air-conditioning Engineers (ASHRAE)” and the “Commission of European Communities (CEC) ventilation guidelines”. Over the last half-century, there has been an increasing understanding of the relative contributions of particulate matter, organic fumes, and microbial contamination to human health issues inside residences, schools, commercial buildings, and other indoor environments [20][70].References
- Borges, J.L.; Weinberger, E. Selected Non-Fictions; Penguin: London, UK, 2000; p. 575. ISBN 0140290117/9780140290110.
- Reed-Scott, J. Preserving Research Collections: A Collaboration between Librarians and Scholars. The Association of Research Libraries, the Modern Language Association, and the American Historical Association on Behalf of the Task Force on the Preservation of the Artifact. 1999. Available online: http://www.arl.org/preserv/prc.html (accessed on 23 June 2023).
- Orlean, S.; Schuster, S. The Library Book; Inner Traditions: Rochester, VT, USA, 2007; p. 336. ISBN 978-1-4767-4018-8.
- Polastron, L.X. Books on Fire; Inner Traditions: Rochester, VT, USA, 2007; p. 384. ISBN 9781594771675.
- Hammer, J. The Bad-Ass Librarians of Timbuktu; Simon & Schuster: New York, NY, USA, 2017; p. 388. ISBN 9781476777412.
- Zyska, B.; Żakowska, Z. Materials Microbiology; Lodz University of Technology: Łódź, Poland, 2005.
- Florian, E.M.L. The role of the conidia of fungi in fox spots. Stud. Conserv. 1996, 41, 65–75.
- Ogden, B. Collection Preservation in Library Building Design; Libris Design Project; The U.S. Institute of Museum and Library Services: Washington, DC, USA, 2004; Available online: https://connectingtocollections.org/collection-preservation-in-library-building-design/ (accessed on 1 May 2023).
- Burge, H.A.; Person, D.L.; Groves, T.O.; Strawn, K.E.; Mishra, S.K. Dynamics of airborne fungal populations in a large office building. Curr. Microbiol. 2000, 40, 10–16.
- Burge, H. An update on pollen and fungal spore aerobiology. J. Allergy Clin. Immunol. 2002, 110, 544–552.
- Florian, M.; Koestler, R.; Nicholson, K.; Parker, T.; Stanlley, T.; Szczepanowska, H.; Wagner, S. Chapter 12-Mold/fungi. In Paper Conservation Catalog; Bertalan, S., Ed.; American Institute for Conservation/Book and Paper Group: Washington, DC, USA, 1994; pp. 1–39.
- Konkol, N.R.; McNamara, C.J.; Hellman, E.; Mitchell, R. Early detection of fungal biomass on library materials. J. Cult. Herit. 2012, 13, 115–119.
- Rawlinson, S.; Ciric, L.; Cloutman-Green, E. How to carry out microbiological sampling of healthcare environment surfaces? A review of current evidence. J. Hosp. Infect. 2019, 103, 363–374.
- Prezant, B.; Weekes, D.M.; Miller, J.D. (Eds.) Recognition, Evaluation, and Control of Indoor Mold; American Industrial Hygiene Association: Fairfax, VA, USA, 2008.
- Chin, Y.A.; Heinsohn, P.A. Sampling and Analysis of Indoor Microorganisms; Wiley Interscience: Hoboken, NJ, USA, 2007.
- Flannigan, B.; Samson, R.A.; Miller, J.D. Microorganisms in Home and Indoor Work Environments: Diversity, Health Impacts, Investigation and Control; CRC Press: London, UK, 2011.
- Verdier, T.; Coutand, M.; Bertron, A.; Roques, C. A review of indoor microbial growth across building materials and sampling and analysis methods. Build. Environ. 2014, 80, 136–149.
- Martin-Sanchez, P.M.; Nunez, M.; Estensmo, E.L.F.; Skrede, I.; Kauserud, H. Comparison of methods to iIdentify and monitor mold damages in buildings. Appl. Sci. 2022, 12, 9372.
- Reponen, T. Sampling for microbial determinations. In Exposure to Microbiological Agents in Indoor and Occupational Environments; Viegas, C., Viegas, S., Gomes, A., Täubel, M., Sabino, R., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 85–96.
- EPA. Collection of Air Samples Potentially Contaminated with Microbiological Agents Using Impingers, Impactors, and Low-Volume Filters; EPA: Cincinnati, OH, USA, 2021.
- Rapid Microbiology. Air Samplers for Microbiological Monitoring of Air Quality. Available online: https://www.rapidmicrobiology.com/test-method/air-samplers (accessed on 22 September 2023).
- Ghosh, B.; Lal, H.; Srivastava, A. Review of bioaerosols in indoor environment with special reference to sampling, analysis and control mechanisms. Environ. Int. 2015, 85, 254–272.
- Lin, X.J.; Reponen, T.; Willeke, K.; Wang, Z.; Grinshpun, S.A.; Trunov, M. Survival of airborne microorganisms during swirling aerosol collection. Aerosol. Sci. Technol. 2000, 32, 184–196.
- Chen, Y.C.; Wang, I.J.; Cheng, C.C.; Wu, Y.C.; Bai, C.H.; Yu, K.P. Effect of selected sampling media, flow rate, and time on the sampling efficiency of a liquid impinger packed with glass beads for the collection of airborne viruses. Aerobiologia 2021, 37, 243–252.
- Burge, H.A. Bioaerosols, 1st ed.; CRC Press: Boca Raton, FL, USA, 1995.
- Andersen, G.L.; Frisch, A.S.; Kellogg, C.A.; Levetin, E.; Lighthart, B.; Paterno, D. Aeromicrobiology/Air Quality. Encycl. Microbiol. 2009, 11–26.
- Wu, F.Q. Culture-based analytical methods for investigation of indoor fungi. In Sampling and Analysis of Indoor Microorganisms; Yang, C.S., Heinsohn, P.A., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2007.
- Kastango, E.S.; Faylor, K. The Importance of Environmental Monitoring, Part II: Surface Testing. 2005, 2, 5. Available online: https://www.pppmag.com/article/100/September_2005/The_Importance_of_Environmental_Monitoring_Part_II_Surface_Testing/ (accessed on 13 September 2023).
- Frankel, M.; Bekö, G.; Timm, M.; Gustavsen, S.; Hansen, E.W.; Madsen, A.M. Seasonal variations of indoor microbial exposures and their relation to temperature, relative humidity, and air exchange rate. Appl. Environ. Microbiol. 2012, 78, 8289–8297.
- Macher, J. (Ed.) Bioaerosol Assessment and Control; America Conference of Government Industrial Hygienists: Cincinnati, OH, USA, 1999.
- Buttner, M.P.; Stetzenbach, L.D. Monitoring airborne fungal spores in an experimental indoor environment to evaluate sampling methods and the effects of human activity on air sampling. Appl. Environ. Microhiol. 1993, 59, 219–226.
- Flannigan, B. Air sampling for fungi in indoor environments. J. Aerosol. Sci. 1997, 28, 381–392.
- Zhou, G.; Whong, W.Z.; Ong, T.; Chen, B. Development of a fungus-specific PCR assay for detecting low-level fungi in an indoor environment. Mol. Cell Probes 2000, 14, 339–348.
- Jeewon, R.; Hyde, K.D. Detection and diversity of fungi from environmental samples: Traditional versus molecular approaches. In Advanced Techniques in Soil Microbiology. Soil Biology; Varma, A., Oelmüller, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 11.
- Niemeier, R.T.; Sivasubramani, S.K.; Reponen, T.; Grinshpun, S.A. Assessment of fungal contamination in moldy homes: Comparison of different methods. J. Occup. Environ. Hyg. 2006, 3, 262–273.
- Nazaroff, W.W. Indoor bioaerosol dynamics. Indoor Air 2014, 26, 61–78.
- David Miller, J.; Christopher Young, J. The Use of Ergosterol to Measure Exposure to Fungal Propagules in Indoor Air. Am. Ind. Hyg. Assoc. J. 1997, 58, 39–43.
- Florian, M.L.E. Fungal-problem monitoring for heritage collections: The need for baseline reference levels for fungal structures and beta-glucans. In Art, Biology, and Conservation: Biodeterioration of Works of Art; Metropolitan Museum of Art: New York, NY, USA, 2003; pp. 172–187.
- Michaelsen, A.; Pinzari, F.; Ripka, K.; Lubitz, W.; Piñar, G. Application of molecular techniques for identification of fungal communities colonising paper material. Int. Biodeterior. Biodegrad. 2006, 58, 133–141.
- Canhoto, O.; Pinzari, F.; Fanelli, C.; Magan, N. Application of electronic nose technology for the detection of fungal contamination in library paper. Int. Biodeterior. Biodegrad. 2004, 54, 303–309.
- Green, B.J.; Yli-Panula, E.; Tovey, E.R. Halogen Immunoassay, a New Method for the Detection of Sensitization to Fungal Allergens; Comparisons with Conventional Techniques. Allergol. Int. 2006, 55, 131–139.
- Sangeetha, J.; Thangadurai, D. Staining Techniques and Biochemical Methods for the Identification of Fungi. In Laboratory Protocols in Fungal Biology. Fungal Biology; Gupta, V., Tuohy, M., Ayyachamy, M., Turner, K., O’Donovan, A., Eds.; Springer: New York, NY, USA, 2013.
- Cox, J.; Indugula, R.; Vesper, S.; Zhu, Z.; Jandarov, R.; Reponen, T. Comparison of indoor air sampling and dust collection methods for fungal exposure assessment using quantitative PCR. Environ. Sci. Process. Impacts 2017, 19, 1312–1319.
- Xu, J. Fungal DNA barcoding. Genome 2016, 59, 913–932.
- Amend, A.S.; Seifert, K.A.; Samson, R.; Bruns, T.D. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc. Natl. Acad. Sci. USA 2010, 107, 13748–13753.
- Korpelainen, H.; Pietiläinen, M.; Huotari, T. Effective detection of indoor fungi by metabarcoding. Ann. Microbiol. 2016, 66, 495–498.
- Schena, L.; Nigro, F.; Ippolito, A.; Gallitelli, D. Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. Eur. J. Plant Pathol. 2004, 110, 893–908.
- Langsiri, N.; Worasilchai, N.; Irinyi, L.; Jenjaroenpun, P.; Wongsurawat, T.; Luangsa-Ard, J.J.; Meyer, W.; Chindamporn, A. Targeted sequencing analysis pipeline for species identification of human pathogenic fungi using long-read nanopore sequencing. IMA Fungus 2023, 14, 18.
- Portnoy, J.M.; Barnes, C.S.; Kennedy, K. Sampling for indoor fungi. J. Allergy Clin. Immunol. 2004, 113, 189–198.
- Dannemiller, K.C.; Lang-Yona, N.; Yamamoto, N.; Rudich, Y.; Peccia, J. Combining real-time PCR and next-generation DNA sequencing to provide quantitative comparisons of fungal aerosol populations. Atmos. Environ. 2014, 84, 113–121.
- Stengel, A.; Stanke, K.M.; Quattrone, A.C.; Herr, J.R. Improving taxonomic delimitation of fungal species in the age of genomics and Phenomics. Front. Microbiol. 2022, 13, 847067.
- Rakotonirainy, M.S.; Bénaud, O.; Vilmont, L.-B. Contribution to the characterization of foxing stains on printed books using infrared spectroscopy and scanning electron microscopy energy dispersive spectrometry. Int. Biodeterior. Biodegrad. 2015, 101, 1–7.
- Lecellier, A.; Gaydou, V.; Mounier, J.; Hermet, A.; Castrec, L.; Barbier, G.; Ablain, W.; Manfait, M.; Toubas, D.; Sockalingum, G.D. Implementation of an FTIR spectral library of 486 filamentous fungi strains for rapid identification of molds. Food Microbiol. 2015, 45, 126–134.
- Barboux, R.; Bousta, F.; Di Martino, P. FTIR Spectroscopy for Identification and Intra-Species Characterization of Serpula lacrymans. Appl. Sci. 2021, 11, 8463.
- Inamdar, A.A.; Morath, S.; Bennett, J.W. Fungal Volatile Organic Compounds: More Than Just a Funky Smell? Annu. Rev. Microbiol. 2020, 74, 101–116.
- Schuchardt, S.; Kruse, H. Quantitative volatile metabolite profiling of common indoor fungi: Relevancy for indoor air analysis. J. Basic Microbiol. 2009, 49, 350–362.
- Cincinelli, A.; Martellini, T.; Amore, A.; Dei, L.; Marrazza, G.; Carretti, E.; Belosi, F.; Ravegnani, F.; Leva, P. Measurement of volatile organic compounds (VOCs) in libraries and archives in Florence (Italy). Sci. Total Environ. 2016, 572, 333–339.
- Gibson, L.T.; Ewlad-Ahmed, A.; Knight, B.; Horie, V.; Mitchell, G.; Robertson, C.J. Measurement of volatile organic compounds emitted in libraries and archives: An inferential indicator of paper decay? Chem. Cent. J. 2012, 6, 42.
- Fantuzzi, G.; Aggazzotti, G.; Righi, E.; Cavazzuti, L.; Predieri, G.; Franceschelli, A. Indoor air quality in the university libraries of Modena (Italy). Sci. Total Environ. 1996, 193, 49–56.
- RTI Laboratories. SEM/EDS Analysis. Available online: https://rtilab.com/techniques/sem-eds-analysis/ (accessed on 22 September 2023).
- Williams, S.T.; Veldkamp, C.J. Preparation of fungi for scanning electron microscopy. Trans. Br. Mycol. Soc. 1974, 63, 408–412.
- Orrance, J.S. A justification of air-conditioning in libraries. J. Librariansh. 1975, 7, 199–206.
- Shum, C.; Alipouri, Y.; Zhong, L. Examination of human interaction on indoor environmental quality variables: A case study of libraries at the University of Alberta. Build. Environ. 2022, 207, 108476.
- Preston, L. Mold Prevention and Remediation in a Library Environment. Tex. Libr. J. 2015, 91, 60.
- René, T. Preservation of Archives in Tropical Climates: An Annotated Bibliography; International Council on Archives: Paris, France; National Archives of the Netherlands: The Hague, The Netherlands; National Archives of the Republic of Indonesia: Daerah Khusus Ibukota Jakarta, Indonesia, 2001.
- Wood, L. Prevention and Treatment of Molds in Library Collections, Especially in Tropical Climates: A RAMP Study; Prepared by Mary Wood Lee General Information Program and UNISIST; UNESCO: Paris, France, 1988; 56p.
- Ren, P.; Jankun, T.M.; Belanger, K.; Bracken, M.B.; Leaderer, B.P. The relation between fungal propagules in indoor air and home characteristics. Allergy 2001, 56, 419–424.
- Bamba, I.; Azuma, M.; Hamada, N.; Kubo, H.; Isoda, N. Case study of airborne fungi according to air temperature and relative humidity in houses with semibasements adjacent to a forested hillside. Biocontrol Sci. 2014, 19, 1–9.
- Brown, D.R. Collection disaster: Mold in the stacks. Coll. Res. Libr. News 2003, 64, 304–306.
- National Academies of Sciences, Engineering, and Medicine. Microbiomes of the Built Environment: A Research Agenda for Indoor Microbiology, Human Health, and Buildings; The National Academies Press: Washington, DC, USA, 2017.
More
