The endogenous miRNAs of breast milk are the products of more than 1000 nonprotein-coding genes, giving rise to mature small regulatory molecules of 19–25 nucleotides. They are incorporated in macromolecular complexes, loaded on Argonaute proteins, sequestrated in exosomes and lipid complexes, or present in exfoliated cells of epithelial, endothelial, or immune origins. Their expression is dependent on the stage of lactation; however, their detection depends on progress in RNA sequencing and the reappraisal of the definition of small RNAs. Some miRNAs from plants are detected in breast milk, opening the possibility of the stimulation of immune cells from the allergy repertoire. Each miRNA harbors a seeding sequence, which targets mRNAs, gene promoters, or long noncoding RNAs. Their activities depend on their bioavailability. Efficient doses of miRNAs are estimated to be roughly 100 molecules in the cytoplasm of target cells from in vitro and in vivo experiments. Each miRNA is included in networks of stimulation/inhibition/sequestration, driving the expression of cellular phenotypes. Three types of stress applied during lactation to manipulate miRNA supply were explored using rodent offspring: a foster mother, a cafeteria diet, and early weaning.
- nutritional programming
- neonate
- miR-26
- miR-320
1. Introduction
2. RNA Content and Mature miRNA Diversity
The miRNA composition of breast milk has been explored by RNA sequencing performed with kits designed with an RNA extraction step [14] or without [15], as in a recent breast milk cohort [16]. q-PCR is used for the specific exploration of known miRNAs and confirmation studies, with normalization provided by reference endogenous genes like miRNAs or by spiking samples with xenogenous miRNA like cel-lin4-5p. In clinical practice, breast milk is classified as a noninvasive sample available in maternity wards or collected at home on a worldwide scale. The main parameters to ponder when designing a clinical plan are the mother’s delivery (term or preterm), the capacity of mammary glands to deliver the food (difference between breasts; the compositional variation during suckling between fore, middle, and hind milk; and the circadian rhythm of production) (Figure 1A). To control for the differences between breasts, mothers are sometimes asked to use the same breast for sampling at each time-point. Minimal differences in miRNA content have been found between fore and hind milk [17]. It has been advised that pre-feed samples should be utilized in order to minimize confounding [18]. Minimal impacts of freeze–thaw cycles have been found on milk miRNAs [19[19][20],20], which is not surprising as the milk of domestic animals retains cryoprotective properties. All these parameters have to be taken into account when designing a sampling procedure [21]. In clinical trials, available volumes varying between 100 µL and 100 mL have been reported (Figure 1B). Although q-PCR is a sensitive method, the design of a breast milk analysis using 100 µL (down to 50 µL [20]) is challenging, but it is a prerequisite when exploring milk samples from the same mother during a nychthemeron or when sharing samples between multiple analyses (transcriptomic, lipidomic, and metabolomic [22]). We have to consider that the sampling of a single drop of breast milk clearly has less informative value than a volume of over 5 mL. Multiplexed techniques to allow bulk extractions from low volumes of crude milk are needed in the field. The RNA content of whole breast milk after classical phenol/chloroform extraction is wide; for instance, 90 to 1000 ng/µL from 8 parturients [23]. Breast milk contains a high amount of small or very small noncoding RNAs [24] along with transfer RNA, messenger RNA, and long noncoding RNA [25,26][25][26].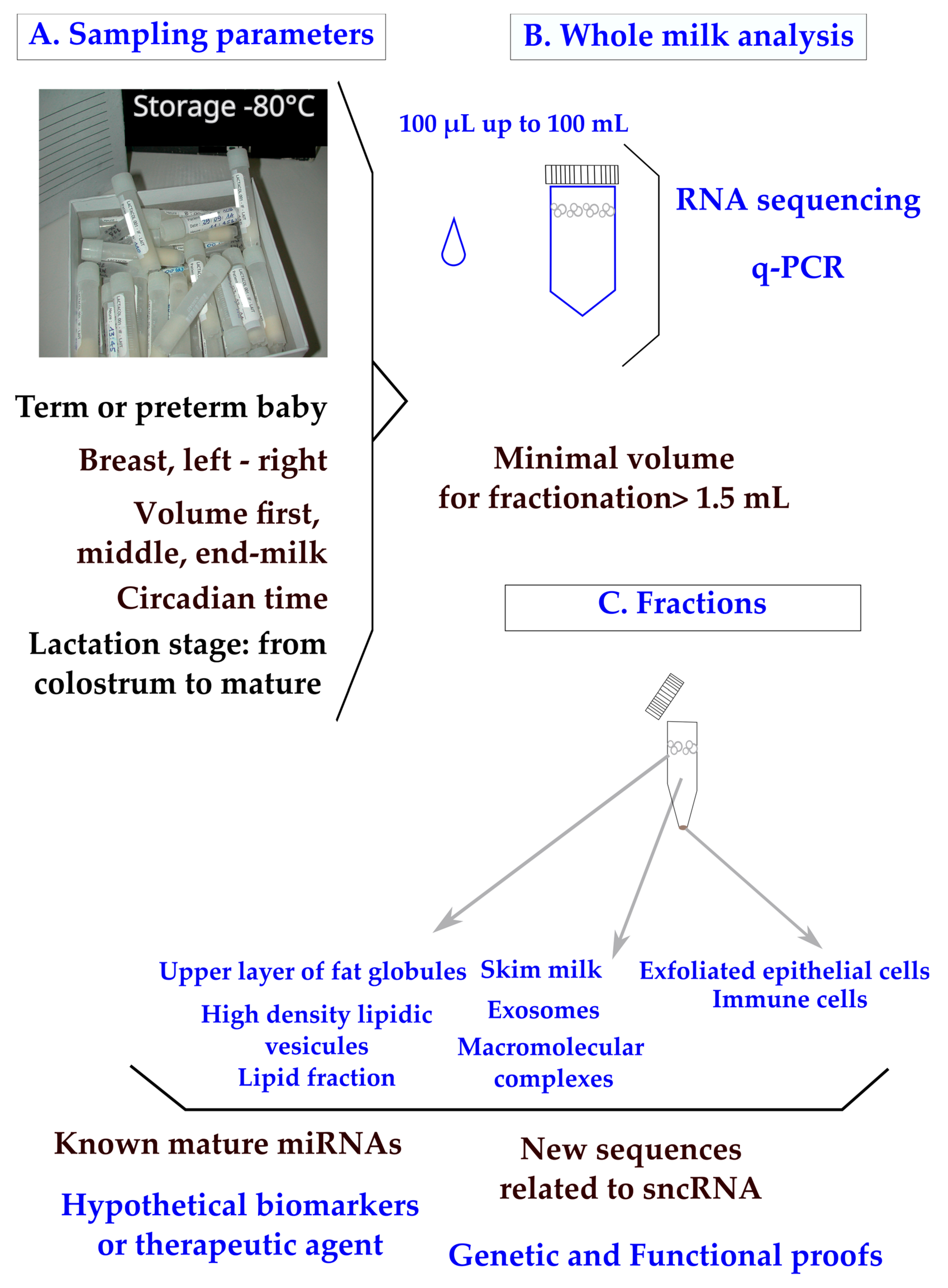
3. Bioavailability of Mature miRNAs in the Digestive System
The systemic RNA interference–deficient transporter (sidt1) is the main receptor of dietary and orally administered miRNAs in the digestive system [53][41], indicating that miRNAs present in breast milk can transfer to offspring. However, miR-375-3p cannot cross the digestive tract of mice [54][42]; even when using the breast milk of a mouse engineered to produce a high amount of miR-30b, a demonstration of plasma loading was impossible [55][43]. As a consequence, the bioavailability of miRNA depends on sheltering these molecules from the molecular environment. It determines the capacity of miRNA molecules to reach target cells with a concentration high enough to trigger a biological response, a crucial step in digestive fluids highly loaded with RNases. In vitro data on cell cultures have shown that a ratio of 100 miRNA molecules delivered to the target cell cytoplasm triggers a measurable physiological response [56][44]. In vivo, after a rough estimation of the total cells of a rat gastric mucosa to adapt to a concentration of miR-320-3p or miR-375-3p, we were able to confirm that this ratio could be used in an oral gavage [57,58][45][46]. Milk is rich in exosomes, which can serve as natural cargo for miRNAs [59][47]. However, these molecules are frequently widely present in all milk fractions. Thus, fluorescently labeled miRNAs like miR-375-3p must be transfected in exosomes in order to show accumulations in the liver, spleen, and brain after suckling or oral gavages. A demonstration was conducted using mice and by transfecting bovine exosomes with fluorescent miRNA administered to mice [60][48]. Another approach is to take advantage of the milk composition related to a certain pathology of the mother. Human milk exosomes from gestational diabetes mellitus (GDM) and healthy parturients showed distinct regulatory bioactivities in both HepG2 cell cultures and, in vivo, in the liver of Balb/c mice. The profile of GDM exosomes has been related to natural loading by miR-101-3p [34]. Previous studies confirmed that the target gene of miR-101-3p is mTOR. miR-101-3p suppresses mTOR by binding in the 3′-UTR regions of mRNAs [61][49]. Works on milk exosomes are a very active field of investigation because (1) a food source of a single miRNA species to supplement diets with a crude product does not exist; (2) exosomes can protect RNA from digestive enzymes; and (3) they can be tailored by the genetic engineering of cell lines to load miRNAs with molecular data, addressing relevant cells or tissues [62][50]. Some natural miRNAs can be addressed to exosomes [63][51] with a step of loading the miRNA onto Argonaute proteins before loading into exosomes [64][52]. However, miRNA obtained by chemical synthesis can also be loaded into artificial vectors like lipoaminoglycoside Dioleyl-Succinyl Paromomycin [65][53]. The loading of miR-375-3p has been measured using a transgenic rat model on the enteroendocrine cell lineage [58][46]. Taking this specific miRNA as an example, miR-375-3p has been proposed as a key regulator in malignant breast cancer [66][54]. In cohorts of breast-fed infants, the consumption of miR-375-3p was associated with protection from atopy [67][55], opening the consideration of the prevention of allergic diseases through breastfeeding. The levels of miR-375-3p were found to be upregulated in the breast milk of mothers treated with probiotics, although the restudyearch concluded that this miRNA and others could not be considered to convey protection against allergic diseases to the baby [68][56]. These divergent properties illustrate the problem caused by supplying miRNA in an animal model; most of the time, the site of delivery impacted different cellular phenotypes. Another level of complexity is added if one considers that the immune cells involved in allergy may be also stimulated by miRNAs from plants present in breast milk [23]. As the chemistry of plant miRNAs is different from eucaryotes, complicating the analysis of breast milk, the presence of these xenogenous molecules has to be taken into account in future allergy studies [69][57]. The oral administration of miRNAs is a current problem in targeted nutritional therapy and requires further studies, both to design new prokaryotic or eukaryotic vectors [70,71][58][59] and to test miRNA cocktails. A specific problem with models of rodent babies is to ensure that the stomach is empty by separating the pups from their mother in one hour before gavage [57,58][45][46]. The biological relevance of miRNAs in low amounts is counterintuitive as a high abundance frequently correlates with a high bioactivity [72][60]. But, the continued uptake of milk-derived exosomes that carry dnmt-targeting miRNAs may promote diabetes, allergies [69][57], neurodegenerative diseases, and cancer later in life [11].References
- Playford, R.J.; Macdonald, C.E.; Johnson, W.S. Colostrum and milk-derived peptide growth factors for the treatment of gastrointestinal disorders. Am. J. Clin. Nutr. 2000, 72, 5–14.
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279.
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242.
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364.
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51.
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672.
- Li, Z.F.; Liang, Y.M.; Lau, P.N.; Shen, W.; Wang, D.K.; Cheung, W.T.; Xue, C.J.; Poon, L.M.; Lam, Y.W. Dynamic Localisation of Mature MicroRNAs in Human Nucleoli is Influenced by Exogenous Genetic Materials. PLoS ONE 2013, 8, e70869.
- Kren, B.T.; Wong, P.Y.-P.; Sarver, A.; Zhang, X.; Zeng, Y.; Steer, C.J. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009, 6, 65–72.
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741.
- Melnik, B.C.; John, S.M.; Schmitz, G. Milk is not just food but most likely a genetic transfection system activating mTORC1 signaling for postnatal growth. Nutr. J. 2013, 12, 103.
- Melnik, B.C.; Schmitz, G. DNA methyltransferase 1-targeting miRNA-148a of dairy milk: A potential bioactive modifier of the human epigenome. Funct. Foods Health Dis. 2017, 7, 671.
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442.
- Melnik, B.C. Milk disrupts p53 and DNMT1, the guardians of the genome: Implications for acne vulgaris and prostate cancer. Nutr. Metab. 2017, 14, 55.
- Zhao, S.; Fung-Leung, W.P.; Bittner, A.; Ngo, K.; Liu, X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells . PLoS ONE. 2014, 9, e78644.
- Songia, P.; Chiesa, M.; Valerio, V.; Moschetta, D.; Myasoedova, V.A.; D’alessandra, Y.; Poggio, P. Direct screening of plasma circulating microRNAs. RNA Biol. 2018, 15, 1268–1272.
- Raymond, F.; Lefebvre, G.; Texari, L.; Pruvost, S.; Metairon, S.; Cottenet, G.; Zollinger, A.; Mateescu, B.; Billeaud, C.; Picaud, J.-C.; et al. Longitudinal Human Milk miRNA Composition over the First 3 mo of Lactation in a Cohort of Healthy Mothers Delivering Term Infants. J. Nutr. 2022, 152, 94–106.
- Hicks, S.D.; Carney, M.C.; Tarasiuk, A.; DiAngelo, S.L.; Birch, L.L.; Paul, I.M. Breastmilk microRNAs are stable throughout feeding and correlate with maternal weight. Transl. Genet. Genom. 2017, 5, 1–8.
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, 6, 20680.
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related MicroRNAs are Abundant in Breast Milk Exosomes. Int. J. Biol. Sci. 2012, 8, 118–123.
- Floris, I.; Billard, H.; Boquien, C.-Y.; Joram-Gauvard, E.; Simon, L.; Legrand, A.; Boscher, C.; Rozé, J.-C.; Bolaños-Jiménez, F.; Kaeffer, B. MiRNA Analysis by Quantitative PCR in Preterm Human Breast Milk Reveals Daily Fluctuations of hsa-miR-16-5p. PLoS ONE 2015, 10, e0140488.
- Holzhausen, E.A.; Kupsco, A.; Chalifour, B.N.; Patterson, W.B.; Schmidt, K.A.; Mokhtari, P.; Baccarelli, A.A.; Goran, M.I.; Alderete, T.L. Influence of technical and maternal-infant factors on the measurement and expression of extracellular miRNA in human milk. Front. Immunol. 2023, 14, 1151870.
- Alexandre-Gouabau, M.-C.; Le Dréan, G.; Kaeffer, B.; Abderrahlan, A.; De Coppet, P.; Bobin, P.; Croyal, M.; De Luca, A.; Hankard, R.; Robitaille, J. Gestational Diabetes Meletis modifies human breast milk content in insulin sensitivity regulators. In Proceedings of the DOHAD, Vancouver, BC, Canada, 27–31 August 2022.
- Lukasik, A.; Brzozowska, I.; Zielenkiewicz, U.; Zielenkiewicz, P. Detection of Plant miRNAs Abundance in Human Breast Milk. Int. J. Mol. Sci. 2017, 19, 37.
- Benmoussa, A.; Laugier, J.; Beauparlant, C.J.; Lambert, M.; Droit, A.; Provost, P. Complexity of the microRNA transcriptome of cow milk and milk-derived extracellular vesicles isolated via differential ultracentrifugation. J. Dairy Sci. 2020, 103, 16–29.
- Karlsson, O.; Rodosthenous, R.S.; Jara, C.; Brennan, K.J.; Wright, R.O.; Baccarelli, A.A.; Wright, R.J. Detection of long non-coding RNAs in human breastmilk extracellular vesicles: Implications for early child development. Epigenetics 2016, 11, 721–729.
- Mourtzi, N.; Siahanidou, T.; Tsifintaris, M.; Karamichali, E.; Tasiopoulou, A.; Sertedaki, A.; Pesmatzoglou, M.; Kapetanaki, A.; Liosis, G.; Baltatzis, G.; et al. lncRNA NORAD is consistently detected in breastmilk exosomes and its expression is downregulated in mothers of preterm infants. Int. J. Mol. Med. 2021, 48, 216.
- Leiferman, A.; Shu, J.; Upadhyaya, B.; Cui, J.; Zempleni, J. Storage of Extracellular Vesicles in Human Milk, and MicroRNA Profiles in Human Milk Exosomes and Infant Formulas. J. Pediatr. Gastroenterol. Nutr. 2019, 69, 235–238.
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk cells and lipids conserve numerous known and novel miRNAs, some of which are differentially expressed during lactation. PLoS ONE 2016, 11, e0152610.
- Alsaweed, M.; Lai, C.T.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human Milk Cells Contain Numerous miRNAs that May Change with Milk Removal and Regulate Multiple Physiological Processes. Int. J. Mol. Sci. 2016, 17, 956.
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H.; et al. Transcriptome Profiling of microRNA by Next-Gen Deep Sequencing Reveals Known and Novel miRNA Species in the Lipid Fraction of Human Breast Milk. PLoS ONE 2013, 8, e50564.
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385.
- Desvignes, T.; Batzel, P.; Berezikov, E.; Eilbeck, K.; Eppig, J.T.; McAndrews, M.S.; Singer, A.; Postlethwait, J.H. miRNA No-menclature: A View Incorporating Genetic Origins, Biosynthetic Pathways, and Sequence Variants. Trends Genet. 2015, 31, 613–626.
- Zonneveld, M.I.; Brisson, A.R.; van Herwijnen, M.J.C.; Tan, S.; van de Lest, C.H.A.; Redegeld, F.A.; Garssen, J.; Wauben, M.H.M.; Nolte-’t Hoen, E.N. Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. J. Extracell. Vesicles 2014, 3, 24215.
- Zheng, Z.; Mo, J.; Lin, F.; Wang, J.; Chen, J.; Luo, H.; Liu, Y.; Su, C.; Gu, X.; Xiong, F.; et al. Milk Exosomes from Gestational Diabetes Mellitus (GDM) and Healthy Parturient Exhibit Differential miRNAs Profiles and Distinct Regulatory Bioactivity on Hepatocyte Proliferation. Mol. Nutr. Food Res. 2023, 67, e2300005.
- Wu, F.; Zhi, X.; Xu, R.; Liang, Z.; Wang, F.; Li, X.; Li, Y.; Sun, B. Exploration of microRNA profiles in human colostrum. Ann. Transl. Med. 2020, 8, 1170.
- Ishibashi, O.; Ohkuchi, A.; Ali, M.; Kurashina, R.; Luo, S.-S.; Ishikawa, T.; Takizawa, T.; Hirashima, C.; Takahashi, K.; Migita, M.; et al. Hydroxysteroid (17-β) Dehydrogenase 1 Is Dysregulated by Mir-210 and Mir-518c That Are Aberrantly Expressed in Preeclamptic Placentas. Hypertension 2012, 59, 265–273.
- Patuleia, S.I.S.; van Gils, C.H.; Cao, A.M.O.; Bakker, M.F.; van Diest, P.J.; van der Wall, E.; Moelans, C.B. The Physiological MicroRNA Landscape in Nipple Aspirate Fluid: Differences and Similarities with Breast Tissue, Breast Milk, Plasma and Serum. Int. J. Mol. Sci. 2020, 21, 8466.
- Layne, T.R.; Green, R.A.; Lewis, C.A.; Nogales, F.; Cruz, T.C.D.; Zehner, Z.E.; Seashols-Williams, S.J. micro RNA Detection in Blood, Urine, Semen, and Saliva Stains After Compromising Treatments. J. Forensic Sci. 2019, 64, 1831–1837.
- Mirza, A.H.; Kaur, S.; Nielsen, L.B.; Størling, J.; Yarani, R.; Roursgaard, M.; Mathiesen, E.R.; Damm, P.; Svare, J.; Mortensen, H.B.; et al. Breast Milk-Derived Extracellular Vesicles Enriched in Exosomes From Mothers With Type 1 Diabetes Contain Aberrant Levels of microRNAs. Front. Immunol. 2019, 10, 2543.
- Karbiener, M.; Pisani, D.F.; Frontini, A.; Oberreiter, L.M.; Lang, E.; Vegiopoulos, A.; Mössenböck, K.; Bernhardt, G.A.; Mayr, T.; Hildner, F.; et al. MicroRNA-26 Family Is Required for Human Adipogenesis and Drives Characteristics of Brown Adipocytes. Stem Cells 2013, 32, 1578–1590.
- Chen, Q.; Zhang, F.; Dong, L.; Wu, H.; Xu, J.; Li, H.; Wang, J.; Zhou, Z.; Liu, C.; Wang, Y.; et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2020, 31, 247–258.
- Title, A.C.; Denzler, R.; Stoffel, M. Uptake and Function Studies of Maternal Milk-derived MicroRNAs. J. Biol. Chem. 2015, 290, 23680–23691.
- Laubier, J.; Castille, J.; Le Guillou, S.; Le Provost, F. No effect of an elevated miR-30b level in mouse milk on its level in pup tissues. RNA Biol. 2015, 12, 26–29.
- Mullokandov, G.; Baccarini, A.; Ruzo, A.; Jayaprakash, A.D.; Tung, N.; Israelow, B.; Evans, M.J.; Sachidanandam, R.; Brown, B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 2012, 9, 840–846.
- Beuzelin, D.; Pitard, B.; Kaeffer, B. Oral Delivery of miRNA With Lipidic Aminoglycoside Derivatives in the Breastfed Rat. Front. Physiol. 2019, 10, 1037.
- Tavares, G.A.; Torres, A.; Le Drean, G.; Queignec, M.; Castellano, B.; Tesson, L.; Remy, S.; Anegon, I.; Pitard, B.; Kaeffer, B. Oral Delivery of miR-320-3p with Lipidic Aminoglycoside Derivatives at Mid-Lactation Alters miR-320-3p Endogenous Levels in the Gut and Brain of Adult Rats According to Early or Regular Weaning. Int. J. Mol. Sci. 2022, 24, 191.
- Liao, Y.; Du, X.; Yalin, L.; Lönnerdal, B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61, 1700082.
- Manca, S.; Upadhyaya, B.; Mutai, E.; Desaulniers, A.T.; Cederberg, R.A.; White, B.R.; Zempleni, J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci. Rep. 2018, 8, 11321.
- Riquelme, I.; Tapia, O.; Leal, P.; Sandoval, A.; Varga, M.G.; Letelier, P.; Buchegger, K.; Bizama, C.; Espinoza, J.A.; Peek, R.M.; et al. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol. 2015, 39, 23–33.
- Horns, F.; Martinez, J.A.; Fan, C.; Haque, M.; Linton, J.M.; Tobin, V.; Santat, L.; Maggiolo, A.O.; Bjorkman, P.J.; Lois, C.; et al. Engineering RNA export for measurement and manipulation of living cells. Cell 2023, 186, 3642–3658.e32.
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2021, 601, 446–451.
- Ghoshal, B.; Bertrand, E.; Bhattacharyya, S.N. Non-canonical argonaute loading of extracellular vesicle-derived exogenous single-stranded miRNA in recipient cells. J. Cell Sci. 2021, 134, jcs253914.
- Beuzelin, D.; Kaeffer, B. Exosomes and miRNA-Loaded Biomimetic Nanovehicles, a Focus on Their Potentials Preventing Type-2 Diabetes Linked to Metabolic Syndrome. Front. Immunol. 2018, 9, 2711.
- Liu, J.; Wang, P.; Zhang, P.; Zhang, X.; Du, H.; Liu, Q.; Huang, B.; Qian, C.; Zhang, S.; Zhu, W.; et al. An integrative bioinformatics analysis identified miR-375 as a candidate key regulator of malignant breast cancer. J. Appl. Genet. 2019, 60, 335–346.
- Hicks, S.D.; Beheshti, R.; Chandran, D.; Warren, K.; Confair, A. Infant consumption of microRNA miR-375 in human milk lipids is associated with protection from atopy. Am. J. Clin. Nutr. 2022, 116, 1654–1662.
- Simpson, M.R.; Brede, G.; Johansen, J.; Johnsen, R.; Storrø, O.; Sætrom, P.; Øien, T. Human breast milk miRNA, maternal probiotic supplementation and atopic dermatitis in offspring. PLoS ONE 2015, 10, e0143496.
- Acevedo, N.; Alashkar Alhamwe, B.; Caraballo, L.; Ding, M.; Ferrante, A.; Garn, H.; Garssen, J.; Hii, C.S.; Irvine, J.; Llinás-Caballero, K.; et al. Perinatal and Early-Life Nutrition, Epigenetics, and Allergy. Nutrients 2021, 13, 724.
- Liu, H.; Geng, Z.; Su, J. Engineered mammalian and bacterial extracellular vesicles as promising nanocarriers for targeted therapy. Extracell. Vesicles Circ. Nucleic Acids 2022, 3, 63–86.
- Afrin, H.; Bai, R.G.; Kumar, R.; Ahmad, S.S.; Agarwal, S.K. Nurunnabi Oral delivery of RNAi for cancer therapy. Cancer Metastasis Rev. 2023, 42, 699–724.
- Gao, F.; Wang, F.; Cao, H.; Chen, Y.; Diao, Y.; Kapranov, P. Evidence for Existence of Multiple Functional Human Small RNAs Derived from Transcripts of Protein-Coding Genes. Int. J. Mol. Sci. 2023, 24, 4163.
