Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Heliana Dundarova and Version 2 by Rita Xu.
Bat lyssaviruses have become the topic of intensive molecular and epidemiological investigations. Since ancient times, rhabdoviruses have caused fatal encephalitis in humans which has led to research into effective strategies for their eradication.
- lyssavirus phylogroups
- Chiroptera
- evolution
1. Introduction
The Order Chiroptera has a Laurasiatherian origin (“laurasian beasts”), evolved between 50 and 70 million years ago (MYA), and has undergone rapid diversification [1][2][1,2]. Due to their capabilities of self-powered flight and echolocation, bats [3] comprise over 20%, or more than 1460 species, of all modern mammals and are globally distributed, with the exception of the extreme polar regions [4]. They have many characteristics that differentiate them from other mammalian species, such as their unique physiology [5][6][5,6], metabolism [7], and immune system [2][8][9][2,8,9]. These features make them a suitable reservoir for viral zoonoses [4][10][11][4,10,11] and more than 200 viruses have been isolated from or detected in bats [12][13][14][12,13,14]. The order comprises 45 species in Europe [15] from two superfamilies, the Rhinolophoidea and Vespertilionoidea [16], representing a natural reservoir of RNA-viruses.
Viruses from 11 families have been isolated on the continent [17] and bat lyssaviruses in Europe (family Rhabdoviridae) have been the subject of detailed reviews [18][19][20][21][18,19,20,21]. Lyssaviruses are a genus of negative-sense single-strand RNA viruses in the family Rhabdoviridae, subfamily Alpharhabdovirinae. Notably, they are members of the order Mononegavirales, which includes other prominent zoonotic pathogens such as filoviruses (Ebola, Marburg, etc.) and the neurotropic Bornaviridae [22]. Based on genetic divergence, lyssaviruses are classified into 21 different viral species. Recently, several putative new lyssaviruses were published [23][24][25][26][23,24,25,26]. Apart from the Mokola virus (MOKV) and Ikoma lyssavirus (IKOV), which have rodents and African civets as a reservoir, respectively [25][27][28][25,27,28], the rest of the lyssaviruses can be transmitted by Chiroptera [27][29][27,29]. According to the most recent ICTV report [24], lyssavirus names are provided here followed by the traditional abbreviations used to identify their isolates: rabies virus (RABV), Aravan virus (ARAV), Australian bat lyssavirus (ABLV), Bokeloh bat lyssavirus (BBLV), Duvenhage virus (DUVV), European bat lyssavirus 1 (EBLV-1), European bat lyssavirus 2 (EBLV-2), Gannoruwa bat lyssavirus (GBLV), Ikoma lyssavirus (IKOV), Irkut virus (IRKV), Khujand virus (KHUV), Lagos bat virus (LBV), Lleida bat lyssavirus (LLEBV), Mokola virus (MOKV), Shimoni bat virus (SHIBV), Kotalahti bat lyssavirus (KBLV), Divača bat lyssavirus (DBLV), West Caucasian bat virus (WCBV), Matlo bat lyssavirus (MBLV), and Lyssavirus Formosa, which includes Taiwan bat lyssavirus 1 (TWBLV-1) and Taiwan bat lyssavirus 2 (TWBLV-2) [21][24][30][31][32][33][34][35][21,24,30,31,32,33,34,35]. In fact, KBLV and MBLV are only tentative lyssaviruses.
2. Origin, Evolution, and Geographic Distribution of Bat Lyssaviruses
Despite the greater diversity of African lyssaviruses [36][55], Hayman et al. [37][56] assumed that they have a Palearctic origin and challenged “Out of Africa” hypothesis. The Lyssaviruses’ most recent common ancestor (MRCA) evolved from an insect rhabdovirus between 7000 and 11,000 years ago [30][38][39][30,57,58] which was transmitted to representatives of the order Chiroptera and spread globally [38][40][57,59]. According to Rupprecht et al. [30], Africa is the most likely home to the ancestors of taxa within the Genus Lyssavirus, family Rhabdoviridae. According to this review, a large number of different lyssaviruses co-evolved with bats as ultimate reservoirs over millions of years. On the other hand, Velasco-Villa et al. [41][60] argue that in the Western Hemisphere before the arrival of the first European colonizers, rabies virus was present only in bats and so-called mesocarnivores (canids, raccoons, skunks, etc.). It is assumed that all mammals are susceptible to infection with the rabies virus. However, it is most possible that lyssaviruses will never be eradicated due to their presence in chiropteran hosts. Lyssaviruses have undergone purifying selection followed by a neutral evolution of the viral genomes [42][61]. The low rate of nonsynonymous evolution of lyssaviruses is probably the result of constraints imposed by the need to replicate in multiple cell types (muscle, peripheral and central nervous systems, and salivary glands) within the host, which in turn boosts cross-species transmission (e.g., different groups of mammals), or because viral proteins are not subject to immune selection, which means existing lyssaviruses are well adapted to their reservoir [43][44][62,63]. The host switching of the classic rabies lyssavirus (RABV) from bats to other mammals is estimated to have occurred 800 to 1400 years ago, which does not explain the timing of the oldest putative human rabies cases, estimated to have circulated 4000 years ago in ancient Mesopotamia [45][46][64,65]. A possible explanation is that the Mesopotamian RABV lineage disappeared as a consequence of genetic drift (loss of polymorphism) or its high fatality rates [45][64]. According to Rupprecht et al. [47][66] and Badrane et al. [48][67], bats are the primary evolutionary host of rabies viruses as a reservoir of all existing lyssaviruses except MOKV and IKOV, whereas other mammals and humans only maintain several lineages of RABV, including the extinct Mesopotamian strain [30][45][49][30,64,68]. In Europe, bat lyssaviruses (Figure 1) were detected in the United Kingdom, the Netherlands, Finland, Denmark, Poland, Czech Republic, Germany, Switzerland, France, Spain, Hungary, Italy, Slovenia, Croatia, Bulgaria, Ukraine, and Russia [19][21][35][50][51][52][53][19,21,35,38,69,70,71]. During the last two decades, previously unknown lyssaviruses were isolated as follows: WCBV in 2002 on the European side of the Caucasus Mts. [54][72], BBLV in 2010 from Germany [55][50], LLEBV in 2011 from Spain [56][73], KBLV in 2017 from Finland [23], and DBLV in 2014 from Slovenia [35].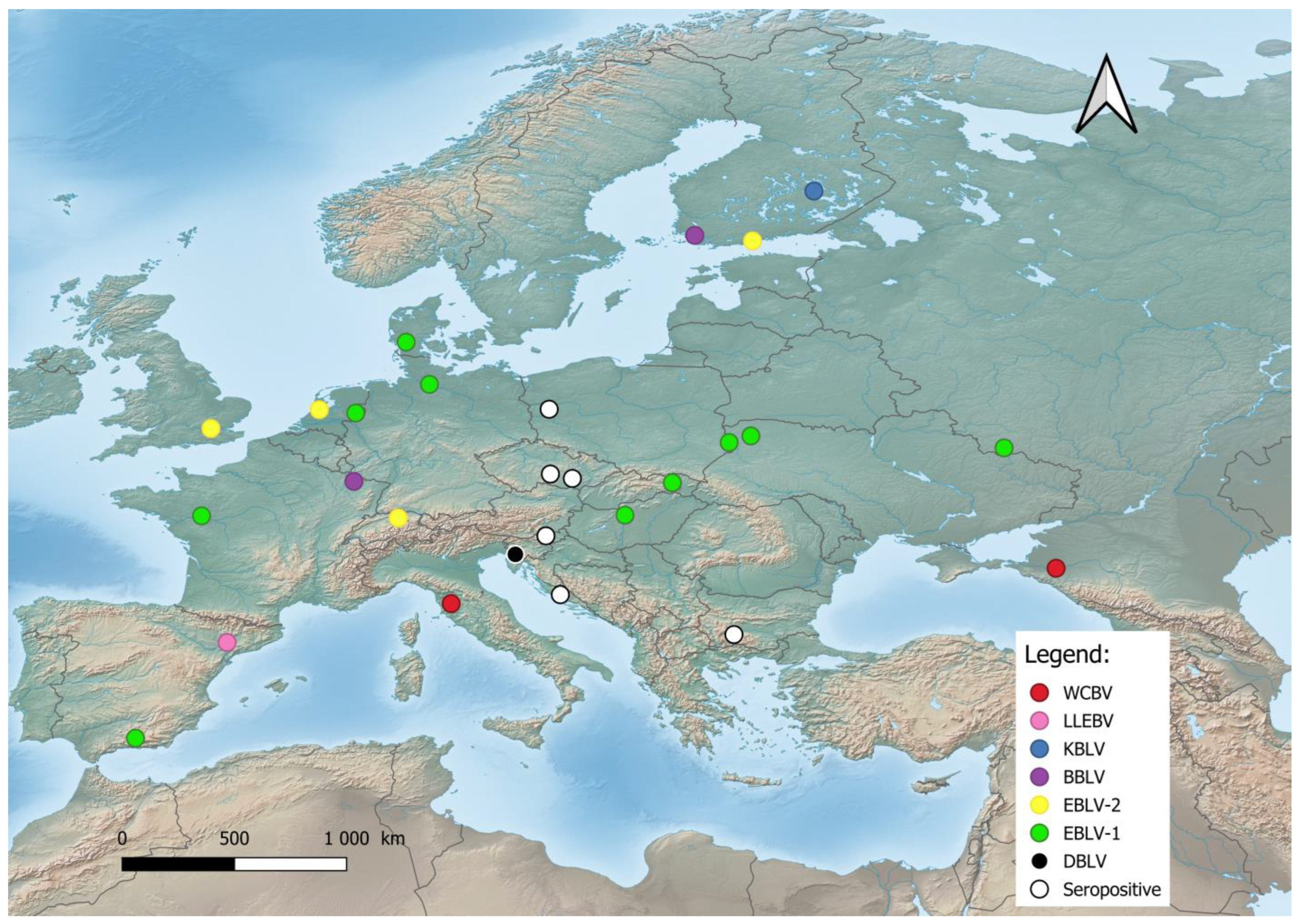

Figure 1. Distribution of bat lyssaviruses in Europe. Abbreviations used: WCBV—West Caucasian bat lyssavirus; LLEBV—Lleida bat lyssavirus; KBLV—Kotalahti bat lyssavirus; BBLV—Bokeloh bat lyssavirus; EBLV-1—European bat lyssavirus 1; EBLV-2—European bat lyssavirus 2; DBLV—Divača bat lyssavirus, Seropositive—Seropositive Blood samples.
3. Phylogeny of Bat Lyssaviruses
Based on the sequence analysis of the lyssavirus N gene, serologic cross-reactivity and pathogenicity bat lyssaviruses are divided into two phylogroups [48][61][62][63][67,80,81,82], https://ictv.global/report/chapter/rhabdoviridae/rhabdoviridae/lyssavirus and an unresolved but widely adopted third phylogroup [64][65][83,84], https://www.who-rabies-bulletin.org/site-page/classification which might contain some of the most divergent lyssaviruses (Figure 2). European viruses are included in Phylogroups I and group of lyssaviruses, which are highly divergent. Phylogroup II is discussed only as a potential scenario for cross-species bat transmission.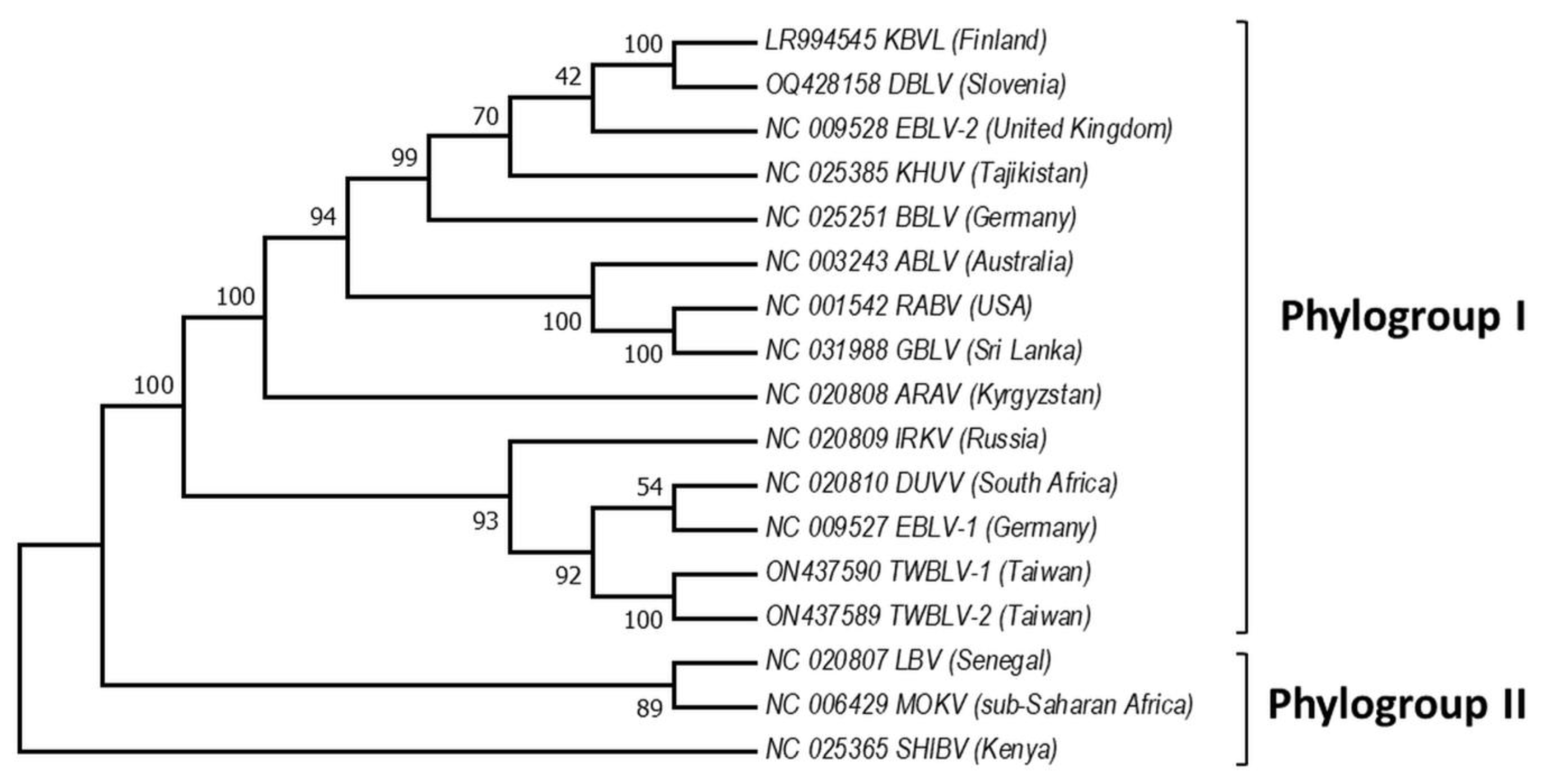

Figure 2. Phylogeny of bat lyssaviruses. The N + P + M + G + L coding regions of representative reference sequences of lyssaviruses used in the analysis were derived from Genbank. The evolutionary history was inferred by using the Maximum Likelihood method and General Time Reversible model. There were a total of 568 positions in the final dataset. Evolutionary analyses were conducted in MEGA X. Virus names are: RABV—rabies virus, ARAV—Aravan virus, ABLV—Australian bat lyssavirus, BBLV—Bokeloh bat lyssavirus, DUVV—Duvenhage virus, EBLV-1—European bat lyssavirus 1, EBLV-2—European bat lyssavirus 2, GBLV—Gannoruwa bat lyssavirus, IKOV—Ikoma virus, IRKV—Irkut virus, KHUV—Khujand virus, LBV—Lagos bat virus, MOKV—Mokola virus, SHIBV—Shimoni bat virus, KBVL—Kotalahti bat lyssavirus, DBLV—Divača bat lyssavirus, TWBLV-1—Taiwan bat lyssavirus 1, and TWBLV-2—Taiwan bat lyssavirus 2.
