Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Lindsay Dong and Version 1 by Dong Han.
Bongkrekic acid (BKA) poisoning, induced by the contamination of Burkholderia gladioli pathovar cocovenenans, has a long-standing history of causing severe outbreaks of foodborne illness. It has emerged as a lethal food safety concern, presenting significant challenges to public health.
- Burkholderia gladioli pathovar cocovenenans
- bongkrekic acid
- food poisoning
- foodborne pathogen
- food safety
1. Introduction
Foodborne diseases have been posing enduring societal and economic challenges worldwide. According to the World Health Organization (WHO), foodborne diseases are estimated to cause 600 million illnesses and 420,000 deaths every year, making them a major cause of morbidity and mortality globally [1,2][1][2]. These illnesses and deaths have a substantial impact on individuals, families, communities, broader society, as well as the global economy. In addition to the human distress caused by these diseases, they also have impactful influences on the food supply chain [2]. Therefore, effective prevention, control, and management of foodborne diseases remain a critical public health priority. Bongkrekic acid (BKA) is a potent respiratory toxin, which could significantly impair mitochondrial ATP/ADP exchange and has been implicated in numerous outbreaks of fatal food poisoning. It is classified as a flavorless, odorless, colorless, thermally stable, and highly unsaturated methoxy tricarboxylic acid [3]. Previous research has demonstrated that BKA is produced by Burkholderia gladioli pv. cocovenenans in natural conditions seeing that the BKA biosynthesis gene cluster (bon) exists in the gene repertoire of this bacterium [4,5,6][4][5][6]. Although the biosynthesis and pathogenesis of BKA are unique in many ways, the bacterium of B. gladioli pv. cocovenenans is prevalently distributed globally as it has been extensively isolated and identified in various food and environmental samples, such as water and soil, from all five most populated continents [7,8][7][8]. Results also indicated that many cultivated food ingredients, such as Tremella fuciformis (white wood ear mushroom), can be contaminated with B. gladioli pv. cocovenenans [9,10][9][10].
2. A Dual Biography of Burkholderia gladioli pathovar cocovenenans and Bongkrekic Acid
At the beginning, B. gladioli pv. cocovenenans was referred to by the name of Pseudomonas cocovenenans [37][11]. In memory of American plant pathologist and microbiologist Walter H. Burkholder, the genus name of Burkholderia was assigned to seven bacterial species that were previously classified within the genus of Pseudomonas [38][12] (Figure 1). After then, this pathogenic bacterium was recognized as Burkholderia cocovenenans, according to genomic comparison study [39][13]. Later, phylogenetic analyses proved that B. cocovenenans is in fact a junior synonym to B. gladioli; therefore, its current nomenclature was conferred [40][14]. While the outbreaks of B. gladioli pv. cocovenenans frequently occurred in coconut-related food consumptions and particularly tempe bongkrèk, a traditional Indonesian fermented coconut food from which BKA derives its name, beforehand, the scientific understanding of this disease remained elusive until the 1930s [24,41][15][16]. Two scientists, van Veen and Mertens, carried out a series of studies on samples from central Java to explore the cause of the then-mysterious deadly food poisoning [42,43,44,45,46][17][18][19][20][21]. There were two poisonous substances discovered from B. gladioli pv. Cocovenenans-contaminated food matrices. Firstly, the researchers identified a yellowish bacterial pigment and named it toxoflavin(e) [44][19]. Toxoflavin is a type of pyrimidotriazine compound that can be produced by multiple Burkholderia spp. (Figure 2A). It has manifested antibiotic, fungicidal, and virulence enhancement functionalities and also exerts certain level toxicities toward eukaryotic cells [47,48,49][22][23][24]. Shortly after, bongkrek(ic) acid was purified and categorized. Although their molecular structures were investigated and proposed after their discovery, the structure of both toxoflavin and BKA was not conclusively determined until the late 1950s [37,50][11][25] (Figure 2B). In contrast to the relatively mild toxicity observed with toxoflavin, BKA exhibits a high level of toxicity in monkeys, with a fatal dose estimated to be approximately 0.5 mg per subject when administered orally to monkeys weighing 1–5 kg [46][21]. Despite evidence that proved that both toxoflavin and BKA were poisonous to human, BKA and its extreme respiratory toxicity have been broadly recognized as the singular cause for the life-threating symptoms given the fact that the toxicity of BKA overwhelmingly outweighs that of toxoflavin in nearly all measurable ways [3,11][3][26].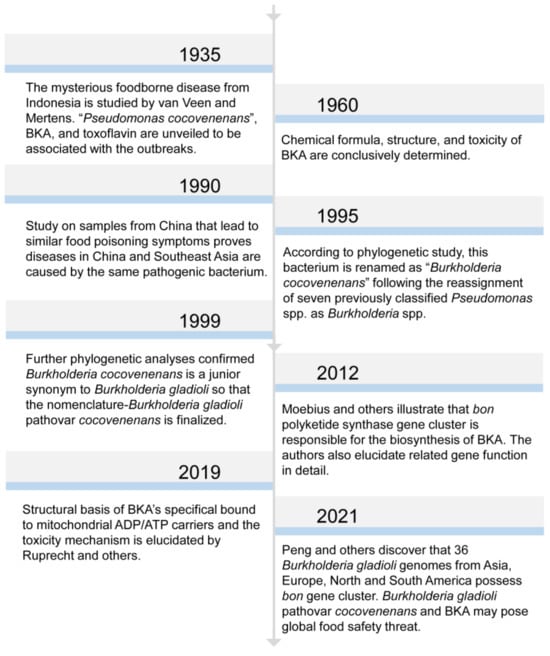
Figure 1. Chronological list of significant research findings in BKA and B. gladioli pv. cocovenenans. The presented years are literature publication years.
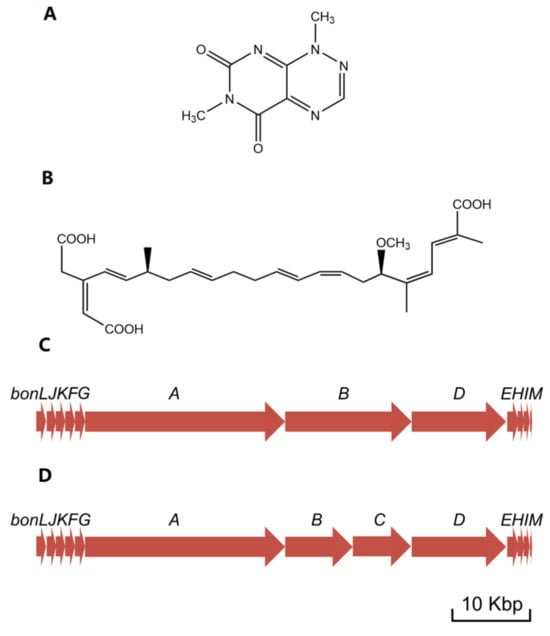
Figure 2. Illustration of related molecular structures and gene clusters. (A) Structure of toxoflavin; (B) Structure of bongkrekic acid; (C) Bongkrekic acid synthesis gene cluster in B. gladioli pv. cocovenenans DSMZ 11318; (D) Bongkrekic acid synthesis gene cluster in B. gladioli pv. cocovenenans BSR3. The presented length of a gene is directly proportional to the quantity of its base pairs.
3. Pathogenesis of Bongkrekic Acid
BKA has been verified to be extremely toxic toward every animal species investigated, such as monkeys, pigeons, rabbit, and rats, even when BKA is administered in non-pure form (fermented coconut cakes) [43][18]. The symptoms observed in animal BKA studies include initial hyperglycemia, subsequent hypoglycemia, and dramatic blood lactic acid content increases (2–3 times of the normal level), and these symptoms were later demonstrated to be associated with impaired mitochondrial oxidative phosphorylation [16][27]. Through respective experimental designs, researchers were able to conclusively determine that BKA could bind to adenine nucleotide translocator (ANT) which located on the mitochondrial inner membrane [24,52,53,54][15][28][29][30].
ANT, also known as ADP/ATP translocase or ADP/ATP carrier, is an important mitochondrial carrier that is responsible for transporting ADP into and ATP out of mitochondria [53,55][29][31]. ANT carriers out its role by cycling between two states: cytoplasmic open state (c-state) and matrix open state (m-state). At normal circumstances, free ADP from mitochondrial intermembrane space (from cell cytoplasm) can specifically bind to c-state ANT, whereas free ADP in mitochondrial matrix can bind to m-state ANT. These binds will lead to conversion cycles between c-state and m-state of ANT and allow the ADP/ATP exchange to be carried out (Figure 3A). When consumed by animals, owing to its apparent lipophilicity, BKA can firstly penetrate the inner mitochondrial membrane and then bind to m-state ANT to form a BKA-inhibited ANT structure (Figure 3B) [13][32].
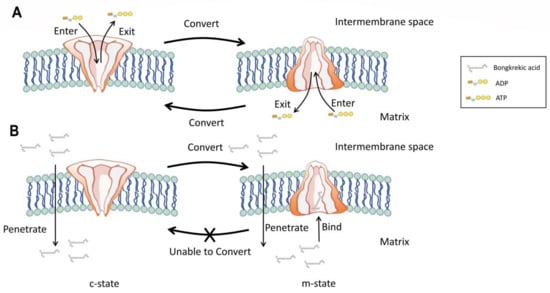

Figure 3. Pathogenic mechanism of BKA. (A) Unimpaired physiological function of mitochondrial ADP/ATP carrier. (B) Function of mitochondrial ADP/ATP carrier impaired by BKA.
4. Genomic Characteristics of Burkholderia gladioli pathovar cocovenenans
4.1. Genomic Diversity and Prevalence
The previous generation of phylogenetic studies on pathogens relies on independent comparisons of live microorganisms or fixed genetic materials, such as cell protein gel electrophoresis, DNA–DNA binding, and detailed biochemical profiling, which was once the only viable method but was deemed as insufficient for both disease control and research purposes [40,56][14][33]. Not long ago, high-throughput sequencing and nucleic-tide-level genome documentation revolutionized this research field swiftly [57,58][34][35]. In silico genomic comparisons among online databases empowered researchers and medical professionals to conduct coherent and efficient genomic epidemiological analyses that were unimaginable previously [59][36]. Genomic assemblies confirmed that B. gladioli pv. cocovenenans incorporates two circular chromosomes with the bon gene cluster located at chromosome 1. It can be concluded from the genome of B. gladioli BSR3 and Co14 that the pathogenic strains also possess one clustered regularly interspaced short palindromic repeat (CRISPR) array sequence at chromosome 1 and a total of five copies of rRNA from the two total chromosomes (Figure 4). A group of researchers [8] conducted species-wide genomic comparative analysis on B. gladioli pv. cocovenenans with the focus on the bon gene cluster (Figure 4).
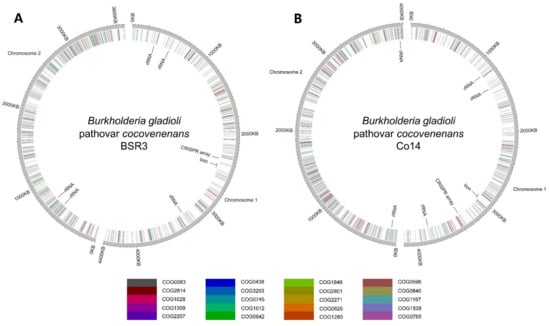

Figure 4. Circular genome diagrams of (A) B. gladioli pv. cocovenenans BSR3 and (B) B. gladioli pv. cocovenenans Co14. Genes of the top 20 abundant clusters of orthologous groups (COGs) are color-labeled according to their COG categories at the respective gene loci. The rRNA gene loci, CRISPR array locus, and bon gene locus are labeled at the inner circle.
Alongside other Burkholderia spp. (both pathogenic and non-pathogenic), a pan-genome analysis was carried out on eight self-assembled genomes of B. gladioli pathovar cocovenenans by another recent research study [5]. The study demonstrated that B. gladioli pv. Cocovenenans exhibits an intricate population structure. Their results also indicated that the ancestor of the pathogenic B. gladioli gained the bon gene cluster (and respective pathogenesis) from horizontal gene transfer. Furthermore, a genome recombination event might cause the deletion of the bon gene cluster from the genome of B. gladioli pv. cocovenenans. This discovery suggested that the ancient B. gladioli may have obtained the bon gene cluster and related regulators from other species, and as they evolved, significant genetic divergence was observed among them.
4.2. The Bon Gene Cluster and Bongkrekic Acid Biosynthesis
Another study [4] employed the Lambda Red homologous recombination technique to establish a bonA-silenced mutant and confirmed that the biosynthesis of BKA is carried out by the bon polyketide synthase (PKS) gene cluster. Using bioinformatic tools, this study then comprehensively depicted the assemble processes and corresponding gene functions in BKA biosynthesis. The center of bon in strain B. gladioli pv. cocovenenans DSMZ 11318 comprises 3 open reading frames (ORFs), bonA, bonB, and bonD (equivalent to 4 ORFs—bonA, bonB, bonC, and bonD—in most other B. gladioli pv. cocovenenans strains) that encode modular type I PKS modules (Figure 3C,D). These PKS modules are essential to BKA biosynthesis since cycles of polyketide chain elongation take place at these modules. The bon gene cluster also comprises nine discrete gene loci, each encoding a free-standing protein to facilitate the biosynthesis of BKA. Specifically, since bon PKS lacks cognate acyltransferase, the loading of an extender unit to acyl carrier protein relies on free-standing cognate acyltransferase BonJ and BonK (Figure 5A) [60,61][37][38].
Similarly, enoyl reductase is absent from the modules in bonA-D, implying that enoyl reductions are carried out by another free-standing protein—BonE (Figure 5B). Researchers also noticed that BKA is rather uncommonly branched compared to many other bacterial PKS products; they deduced that β-branching occurs during the BKA assembly to introduce alkyl branches at the C-21 and C-3 positions in BKA. During the β-branching of BKA, BonF, BonG, and the duo of BonH and BonI most likely function as ketosynthase, 3-Hydroxy-3-methylglutaryl-CoA synthase, and enoyl-CoA hydratase, respectively (Figure 5C) [62][39]. After a typical PKS elongation process, the hydroxyl group at C-17 is methylated by BonM, an O-methyltransferase. Eventually, a novel cytochrome P450 monooxygenase (BonL) was inferred to be responsible for the introduction of a carboxyl group at C-22 (Figure 5D) [4].
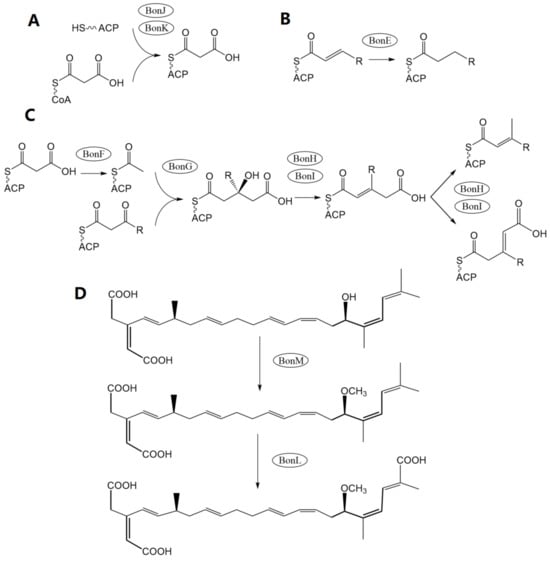

Figure 5. Depiction of essential free-standing gene functions in the biosynthesis of BKA. (A) The loading of the chain-extending unit to acyl carrier protein via cognate acyltransferase BonJ and BonK. (B) Enoyl reductions carried out by enoyl reductase BonE. (C) β-branching carried out by ketosynthase BonF, 3-Hydroxy-3-methylglutaryl-CoA synthase BonG, and enoyl-CoA hydratase BonH and BonI. (D) The terminal synthetic reactions are executed by the catalytic activity of O-methyltransferase BonM and P450 monooxygenase BonL.
5. Characteristics of Burkholderia gladioli pathovar cocovenenans
5.1. Bongkrekic Acid Production in Culture Media
Researchers have conducted a series of investigations into the culture growth and BKA production characteristics of B. gladioli pv. cocovenenans, which have generated valuable insights into the various factors that influence the growth of this bacterium and production of BKA [29,64][40][41]. Their studies have focused on identifying compounds with potential toxin-preventive capabilities to better understand how to control the spread of B. gladioli pv. cocovenenans and reduce the risk of BKA contamination. The study’s significant discovery is that the absence of either coconut oil or glycerol from the culture media completely halts BKA production, without affecting the growth of bacterial populations. This result highlighted the critical role of both coconut oil and glycerol in BKA synthesis and suggested that the fat composition and concentration present in the substrate significantly impacts BKA production. This finding was consistent with the results of another study, which demonstrated that the addition of oleic oil at a concentration of 3.31 mmol per gram of defatted rich coconut medium is a significantly favored substrate for BKA production [9]. The observation indicated that oleic oil, a type of monounsaturated fat that is commonly found in olive oil, might be readily metabolized by B. gladioli pv. cocovenenans for BKA synthesis, resulting in a high level of BKA production. This suggests that specific lipid formation can significantly impact BKA production, with oleic oil being particularly effective.
5.2. Effect of Food Ingredients on the Production of Bongkrekic Acid
In the same study [29[40][41],64], the authors also evaluated the anti-BKA production activities of four natural spices: garlic powder, onion power, capsicum power, and turmeric power. The study found that adding 0.6% garlic powder, 0.6% onion powder, 0.8% capsicum powder, and 0.6% turmeric powder to coconut culture medium could inhibit the formation of BKA when the initial populations of B. gladioli pv. cocovenenans were low (between 3.64 and 5.27 log CFU/mL). However, when the initial populations of B. gladioli pv. cocovenenans were high (above 7.32 log CFU/mL), adding up to 2% of any these spice supplements did not completely inhibit the BKA production.5.3. Bongkrekic Acid Production under Co-Culture Conditions
Beneficial and non-pathogenic microorganisms play a vital role in preventing the development of foodborne pathogens and safeguarding food safety [65][42]. For instance, by occupying the same ecological niches with harmful microorganisms, lactic acid bacteria, such as Lactobacillus spp. and Pediococcus spp., can compete for nutrients and produce antimicrobial substances that inhibit the growth and activity of foodborne pathogens [66][43]. Although present naturally in a diverse range of food commodities, these microbes can also be deliberately introduced or promoted under preferable culture conditions to achieve their dominance in respective food matrices. Moreover, the growth kinetics and antagonistic effects between these microbes and foodborne pathogens, e.g., Salmonella spp., Campylobacter jejuni, Yersinia enterocolitica, pathogenic Escherichia coli, Shigella spp., Vibrio spp., and others, have been previously depicted by employing co-culture experiments [67][44]. However, co-culture studies regarding B. gladioli pv. cocovenenans and BKA production have been rare, considering that the overwhelming majority of BKA outbreaks that occurred in food matrices consist of complex microbial communities. In addition to examining other common factors, the previous study also investigated the impact of Rhizopus oligosporus, the fungus traditionally used for fermenting tempe bongkrèk, on the growth of B. gladioli pv. cocovenenans and the production of BKA under co-culture conditions [29][40].6. Detection and Analytical Advancements
6.1. Detections of Bongkrekic Acid
From the very beginning, the identification and validation of BKA was conducted using paper chromatography methods, which can accurately detect a relatively pure form of BKA at levels as low as 0.05 μg [37,50,71][11][25][45]. Later, with technological advancements, more advanced analytical methods such as HPLC and mass spectrometry (MS) have become available. These instrumental techniques offer enhanced accuracy and sensitivity when compared to paper chromatography, allowing for more efficient and dependable analysis of BKA in both pure formation and in food matrices [33,72][46][47]. The use of the respective methods has facilitated the detection of trace amounts of BKA in complex food matrices, enabling the conduction of highly sophisticated scientific research and reliable epidemiological analyses. With the help of these studies, researchers have acquired a better understanding of BKA in food products [3]. To address the limitations posed by instrumental methods, researchers have turned to alternative techniques that offer greater simplicity, are user-friendly, and are more compatible for in situ food analysis. One major category of these methods is immunoassays, which can quantify BKA based on developed antibodies in theory. The respective immunoassays, such as colloidal gold-based immunochromato-graphic assay (GICA) and enzyme-linked immunosorbent assay (ELISA), have been developed and validated by few Chinese institutions for rapid BKA detection in food products and other types of samples [6,73][6][48]. These methods rely on the robustness of the antibody–antigen reaction to enable efficient and reliable detection of BKA, thus providing a valuable tool for food safety monitoring and quality control.6.2. Detections of Burkholderia gladioli pathovar cocovenenans and Gene
Culture-based analytical methods are crucial for ensuring the accuracy and reliability of the microbial research findings since the culturability of microbes is important to both scientific discovery and prolonged laboratory microbial preservation. Therefore, conventional microbiological analytical approaches, including non-selective medium enrichment, subculturing on differential media, colony observation, microscopic morphology, biochemical tests, gram staining, and toxicity evaluation, have been broadly adopted by research on B. gladioli pv. cocovenenans [6]. The respective analytical protocols were both documented by scientific studies and standardized by government authorities, such as the National Standard of the People’s Republic of China (GB 4789.29–2020) [6,74][6][49]. It Is a justifiable inference that these protocols are labor-intensive, and improvements in these assays could result in substantial benefits in respective studies and regulatory tests. Considering the challenges involved in using traditional culture-based assays, exploring alternative approaches for detecting the presence of B. gladioli pv. cocovenenans could be advantageous. The utilization of novel nucleic-acid-based techniques presents a promising alternative, as they are capable of providing enhanced accuracy and sensitivity in comparison with conventional methods. Therefore, these techniques could be considered as a viable option to identify the presence of B. gladioli pv. cocovenenans [75,76][50][51].7. Conclusions
In summary, the contamination of B. gladioli pv. cocovenenans, leading to the development of bongkrekic acid (BKA) poisoning, has emerged as a critical and lethal food safety concern with dire consequences, posing significant challenges to public health. The incidence of BKA poisoning in recent years has ignited a pursuit for a more profound comprehension of the pathogenicity of B. gladioli pv. cocovenenans and the underlying biochemical mechanisms for BKA synthesis. Specifically, characterization of B. gladioli pv. cocovenenans and the identification of the bon gene cluster have provided an essential foundation for developing targeted interventions to prevent and control BKA accumulation, while high-throughput sequencing and in silico genomic comparisons have unveiled the origin and global prevalence of this pathogen, highlighting this foodborne disease as a significant and ascending threat. Although limited, previous research based on culturing techniques has provided evidence that B. gladioli pv. cocovenenans is capable of producing BKA in different environments, which highlights the possible food safety hazards associated with BKA poisoning. Due to the scarcity of information regarding the impact of culture conditions such as pH, salt content, antimicrobial agents, and coexisting microorganisms on BKA reduction, it is crucial to enhance our understanding of inhibiting BKA production and promote respective applications to ensure food safety. Advancements in the detection methods of both BKA and B. gladioli pv. cocovenenans hold promise for mitigating the impact of this foodborne disease. Overall, it is imperative to undertake future research initiatives on BKA and B. gladioli pv. cocovenenans. Such research endeavors hold significant potential in controlling the threat posed by this formidable adversary to public health.References
- WHO. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; World Health Organization: Geneva, Switzerland, 2015.
- Pires, S.M.; Desta, B.N.; Mughini-Gras, L.; Mmbaga, B.T.; Fayemi, O.E.; Salvador, E.M.; Gobena, T.; Majowicz, S.E.; Hald, T.; Hoejskov, P.S. Burden of foodborne diseases: Think global, act local. Curr. Opin. Food Sci. 2021, 39, 152–159.
- Anwar, M.; Kasper, A.; Steck, A.R.; Schier, J.G. Bongkrekic acid—A review of a lesser-known mitochondrial toxin. J. Med. Toxicol. 2017, 13, 173–179.
- Moebius, N.; Ross, C.; Scherlach, K.; Rohm, B.; Roth, M.; Hertweck, C. Biosynthesis of the respiratory toxin bongkrekic acid in the pathogenic bacterium Burkholderia gladioli. Chem. Biol. 2012, 19, 1164–1174.
- Gong, H.; Huang, X.; Zhu, W.; Chen, J.; Huang, Y.; Zhao, Z.; Weng, J.; Che, Y.; Wang, J.; Wang, X. Pan-genome analysis of the Burkholderia gladioli pv. cocovenenans reveal the extent of variation in the toxigenic gene cluster. Food Microbiol. 2023, 113, 104249.
- Yuan, M.; Han, R.; Bai, L.; Dong, Y.; Xi, Q.; Du, Q.; Yang, Y.; Forghani, F.; Yang, Q.; Ahn, J. Recent Advances in the Characterization of Burkholderia gladioli pv. cocovenenans and Its Toxin Production. Food Rev. Int. 2023, 1–16.
- Falconer, T.M.; Kern, S.E.; Brzezinski, J.L.; Turner, J.A.; Boyd, B.L.; Litzau, J.J. Identification of the potent toxin bongkrekic acid in a traditional African beverage linked to a fatal outbreak. Forensic Sci. Int. 2017, 270, e5–e11.
- Peng, Z.; Dottorini, T.; Hu, Y.; Li, M.; Yan, S.; Fanning, S.; Baker, M.; Xu, J.; Li, F. Comparative genomic analysis of the foodborne pathogen Burkholderia gladioli pv. cocovenenans harboring a bongkrekic acid biosynthesis gene cluster. Front. Microbiol. 2021, 12, 628538.
- Garcia, R.A. The effect of lipids on bongkrekic (Bongkrek) acid toxin production by Burkholderia cocovenenans in coconut media. Food Addit. Contam. 1999, 16, 63–69.
- Zhao, N.; Ma, M.; Zhang, Y.; Xu, D. Comparative description of Pseudomonas cocovenenans (van Damme, Johannes, Cox, and Berends 1960) NCIB 9450T and strains isolated from cases of food poisoning caused by consumption of fermented corn flour in China. Int. J. Syst. Evol. Microbiol. 1990, 40, 452–455.
- Nugteren, D.; Berends, W. Investigations on bongkrekic acid, the toxine from Pseudomonas cocovenenans. Recl. Trav. Chim. PaysBas 1957, 76, 13–27.
- Yabuuchi, E.; Kosako, Y.; Oyaizu, H.; Yano, I.; Hotta, H.; Hashimoto, Y.; Ezaki, T.; Arakawa, M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 1992, 36, 1251–1275.
- Zhao, N.; Qu, C.; Wang, E.; Chen, W. Phylogenetic Evidence for the Transfer of Pseudomonas cocovenenans (van Damme et al. 1960) to the Genus Burkholderia as Burkholderia cocovenenans (van Damme et al. 1960) comb. nov. Int. J. Syst. Evol. Microbiol. 1995, 45, 600–603.
- Coenye, T.; Holmes, B.; Kersters, K.; Govan, J.; Vandamme, P. Burkholderia cocovenenans and Burkholderia vandii are junior synonyms of Burkholderia gladioli (Severini 1913) and Burkholderia plantarii, respectively. Int. J. Syst. Evol. Microbiol. 1999, 49, 37–42.
- Henderson, P.J.; Lardy, H.A. Bongkrekic acid: An inhibitor of the adenine nucleotide translocase of mitochondria. J. Biol. Chem. 1970, 245, 1319–1326.
- Helfrich, E.J.; Piel, J. Biosynthesis of polyketides by trans-AT polyketide synthases. Nat. Prod. Rep. 2016, 33, 231–316.
- Mertens, W.; van Veen, A. A Fréquent Cause of Poisoning in Javanese. Bull. Société Pathol. Exot. 1934, 27. Available online: https://www.cabdirect.org/cabdirect/abstract/19342901639 (accessed on 15 July 2023).
- Van Veen, A.; Mertens, W. On the isolation of a toxic bacterial pigment (provisional communication). Proc. Konink. Akad. Wetensch. Amsterdam 1933, 36, 666–670.
- Van Veen, A.; Mertens, W. Toxoflavine, the yellow pigment of bongkrek. Recl. Trav. Chim. Pays-Bas Belg. 1934, 53, 398–404.
- Van Veen, A.; Mertens, W. Bongkrek acid, a blood sugar lowering substance. Preliminary communication. Recl. Trav. Chim. Pays-Bas Belg. 1935, 54, 373–380.
- Van Veen, A.; Mertens, W. The effect of the acid from bongkrek on carbohydrate metabolism. Geneeskd. Tijdschr. Voor Ned. Indie 1935, 75, 1059–1127.
- Jeong, Y.; Kim, J.; Kim, S.; Kang, Y.; Nagamatsu, T.; Hwang, I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 2003, 87, 890–895.
- Lee, J.; Park, J.; Kim, S.; Park, I.; Seo, Y.S. Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Mol. Plant Pathol. 2016, 17, 65–76.
- Li, X.; Li, Y.; Wang, R.; Wang, Q.; Lu, L. Toxoflavin produced by Burkholderia gladioli from Lycoris aurea is a new broad-spectrum fungicide. Appl. Environ. Microbiol. 2019, 85, e00106-19.
- Van Damme, P.; Johannes, A.; Cox, H.; Berends, W. On toxoflavin, the yellow poison of Pseudomonas cocovenenans. Recl. Trav. Chim. Pays Bas 1960, 79, 255–267.
- Lynch, K.; Dennis, J. Burkholderia; CRC Press: Boca Raton, FL, USA, 2009.
- Welling, W.; Cohen, J.; Berends, W. Disturbance of oxidative phosphorylation by an antibioticum produced by Pseudomonas cocovenenans. Biochem. Pharmacol. 1960, 3, 122–135.
- Erdelt, H.; Weidemann, M.J.; Buchholz, M.; Klingenberg, M. Some principle effects of bongkrekic acid on the binding of adenine nucleotides to mitochondrial membranes. Eur. J. Biochem. 1972, 30, 107–122.
- Klingenberg, M.; Appel, M.; Babel, W.; Aquila, H. The binding of bongkrekate to mitochondria. Eur. J. Biochem. 1983, 131, 647–654.
- Scherer, B.; Klingenberg, M. Demonstration of the relation between the adenine nucleotide carrier and the structural changes of mitochondria as induced by adenosine 5′-diphosphate. Biochemistry 1974, 13, 161–170.
- Kunji, E.R.; Aleksandrova, A.; King, M.S.; Majd, H.; Ashton, V.L.; Cerson, E.; Springett, R.; Kibalchenko, M.; Tavoulari, S.; Crichton, P.G. The transport mechanism of the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 2379–2393.
- Ruprecht, J.J.; King, M.S.; Zögg, T.; Aleksandrova, A.A.; Pardon, E.; Crichton, P.G.; Steyaert, J.; Kunji, E.R. The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 2019, 176, 435–447.e15.
- Behl, A.; Nair, A.; Mohagaonkar, S.; Yadav, P.; Gambhir, K.; Tyagi, N.; Sharma, R.K.; Butola, B.S.; Sharma, N. Threat, challenges, and preparedness for future pandemics: A descriptive review of phylogenetic analysis based predictions. Infect. Genet. Evol. 2022, 98, 105217.
- Baker, S.; Hanage, W.P.; Holt, K.E. Navigating the future of bacterial molecular epidemiology. Curr. Opin. Microbiol. 2010, 13, 640–645.
- Tang, P.; Croxen, M.A.; Hasan, M.R.; Hsiao, W.W.; Hoang, L.M. Infection control in the new age of genomic epidemiology. Am. J. Infect. Control 2017, 45, 170–179.
- Pallen, M.J.; Loman, N.J.; Penn, C.W. High-throughput sequencing and clinical microbiology: Progress, opportunities and challenges. Curr. Opin. Microbiol. 2010, 13, 625–631.
- Chen, H.; Du, L. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 541–557.
- Cheng, Y.Q.; Coughlin, J.M.; Lim, S.K.; Shen, B. Type I polyketide synthases that require discrete acyltransferases. Meth. Enzymol. 2009, 459, 165–186.
- Walker, P.; Weir, A.; Willis, C.; Crump, M. Polyketide β-branching: Diversity, mechanism and selectivity. Nat. Prod. Rep. 2021, 38, 723–756.
- Buckle, K.; Kartadarma, E. Inhibition of bongkrek acid and toxoflavin production in tempe bongkrek containing Pseudomonas cocovenenans. J. Appl. Microbiol. 1990, 68, 571–576.
- Kartadarma, E. Growth of Pseudomonas cocovenenans and Toxin Production in Tempe Bongkrek; University of New South Wales: Sydney, Australia, 1991.
- Fidan, H.; Esatbeyoglu, T.; Simat, V.; Trif, M.; Tabanelli, G.; Kostka, T.; Montanari, C.; Ibrahim, S.A.; Özogul, F. Recent developments of lactic acid bacteria and their metabolites on foodborne pathogens and spoilage bacteria: Facts and gaps. Food Biosci. 2022, 47, 101741.
- Gao, Z.; Daliri, E.B.-M.; Wang, J.; Liu, D.; Chen, S.; Ye, X.; Ding, T. Inhibitory effect of lactic acid bacteria on foodborne pathogens: A review. J. Food Prot. 2019, 82, 441–453.
- Adams, M.R.; Nicolaides, L. Review of the sensitivity of different foodborne pathogens to fermentation. Food Control 1997, 8, 227–239.
- Lumbach, G.; Cox, H.; Berends, W. Elucidation of the chemical structure of bongkrekic acid—I: Isolation, purification and properties of bongkrekic acid. Tetrahedron 1970, 26, 5993–5999.
- Liang, M.; Chen, R.; Xian, Y.; Hu, J.; Hou, X.; Wang, B.; Wu, Y.; Wang, L. Determination of bongkrekic acid and isobongkrekic acid in rice noodles by HPLC-Orbitrap HRMS technology using magnetic halloysite nanotubes. Food Chem. 2021, 344, 128682.
- Hu, J.; Liang, M.; Xian, Y.; Chen, R.; Wang, L.; Hou, X.; Wu, Y. Development and validation of a multianalyte method for quantification of aflatoxins and bongkrekic acid in rice and noodle products using PRiME-UHPLC-MS/MS method. Food Chem. 2022, 395, 133598.
- Cao, X.; Xu, Z.; Su, Y.; Wang, Y.; Lei, H.; Xiao, J. The rapid detection of bongkrekic acid in foods using colloidal gold immunochromatographic assay. J. Chin. Inst. Food Sci. Technol. 2023, 23, 309–318.
- Li, H.-J.; Dong, L.-H.; Chen, G.-F.; Liu, S.-Y.; Yang, J.-Y.; Yang, J.-Y. Establishment of Droplet Digital PCR Assay for Quantitative Detection of Pseudomonas cocovenenans in Foods. Biotechnol. Bull. 2023, 39, 127.
- Mothershed, E.A.; Whitney, A.M. Nucleic acid-based methods for the detection of bacterial pathogens: Present and future considerations for the clinical laboratory. Clin. Chim. Acta 2006, 363, 206–220.
- Huo, B.; Hu, Y.; Gao, Z.; Li, G. Recent advances on functional nucleic acid-based biosensors for detection of food contaminants. Talanta 2021, 222, 121565.
More
