Within the context of biomembranes, the matrix comprises an array of complex molecules, with fatty acids serving as fundamental building blocks. Fatty acids exist typically in two forms: saturated and unsaturated monocarboxylic acids, whereas both are characterized by a terminal carboxyl (-COOH) group and a terminal methyl (-CH3) group designated as carbon 1 (Δ) and omega (ω or n), respectively. Over the past century, numerous nomenclature systems have been proposed, including trivial, systematic, ∆x, n − x, and lipid numbers. The trivial nomenclature, though prevalent, lacks systematic patterns. In contrast, the systematic nomenclature adheres to a more regular and structured approach, based on the nomenclature of parent hydrocarbons. It involves adding the suffix “oic” to the hydrocarbon name after removing the terminal “e”. This nomenclature also encompasses the identification of the position of the first double bond from the (n), with the series of fatty acids being named accordingly (e.g., n-3, n-6, n-7, and n-9 series). These distinctions among n-fatty acids lead to variations in their properties, consequently influencing the structure and function of biomembranes.
- membranes
- phospholipids
- sphingolipids
- fatty acid
- de novo synthesis
- desaturation
1. Synthesis of Fatty Acids
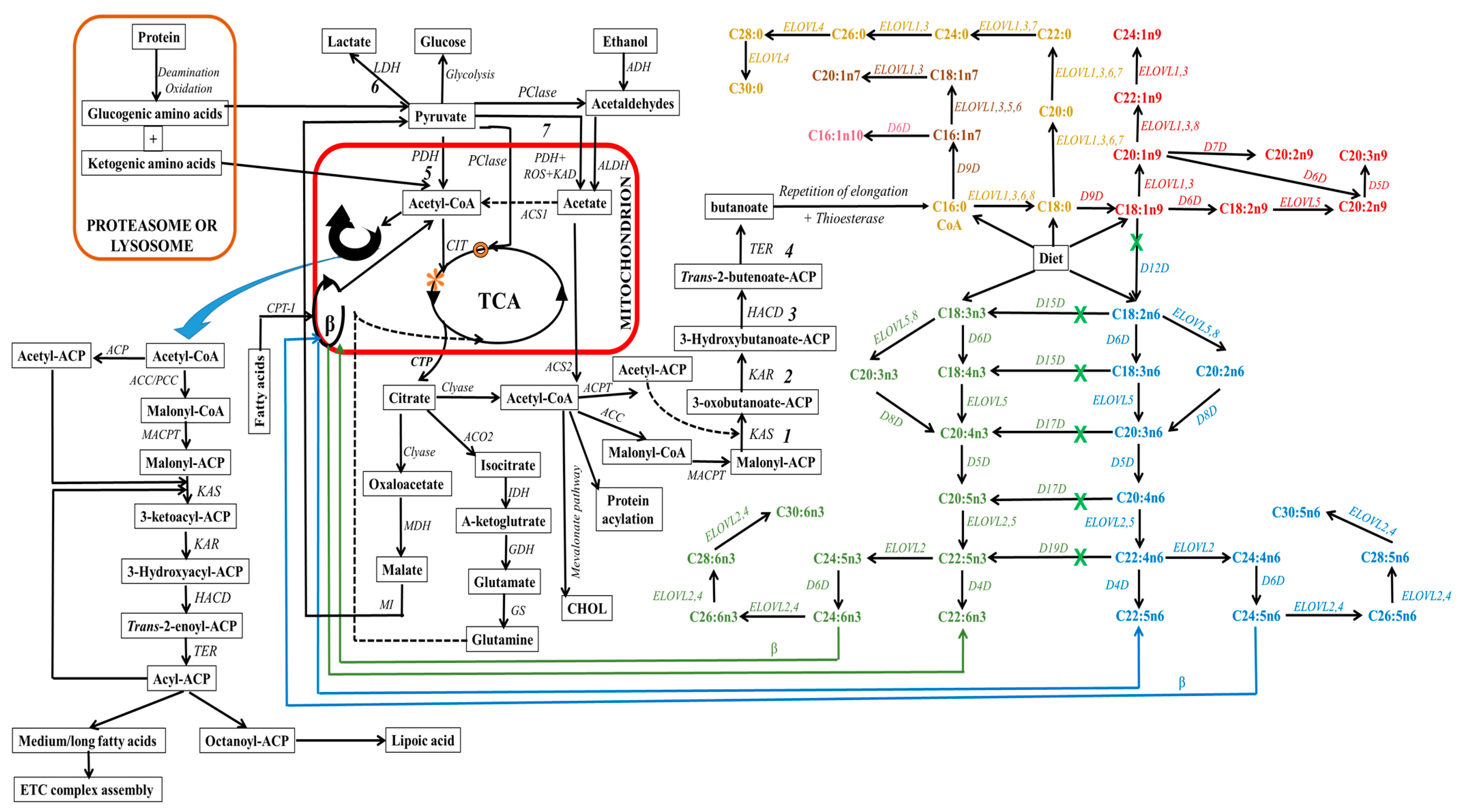
1.1. Elongating Fatty Acids through Non-Cytosolic Mechanisms
1.2. Desaturation of Fatty Acids
2. Incorporation of Fatty Acids into Lipids of Biomembranes
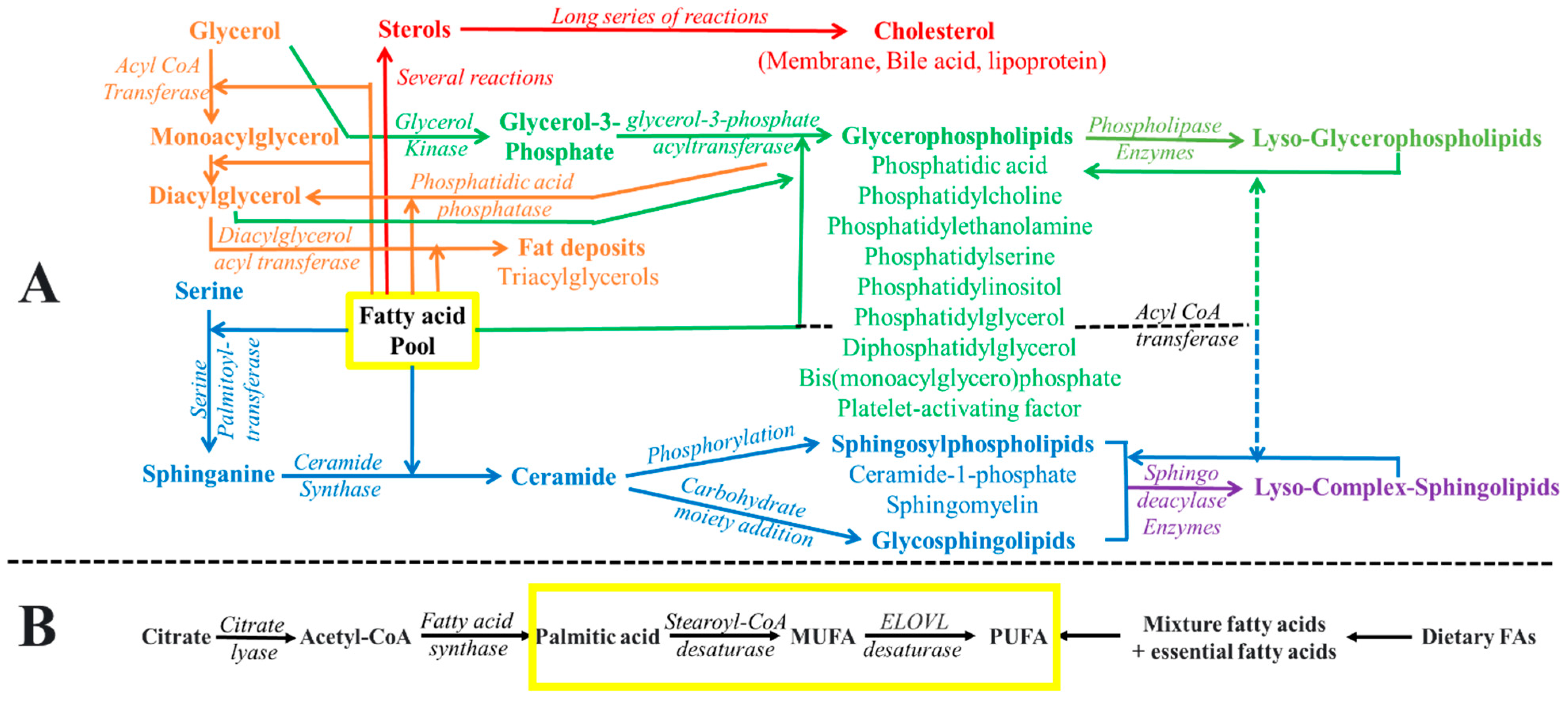
3. Fatty Acid Composition in Biomembranes
3.1. Fatty Acid Profile of Phospholipids
3.2. Fatty Acid Profile of Sphingolipids
References
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532.
- Jalil, A.; Bourgeois, T.; Ménégaut, L.; Lagrost, L.; Thomas, C.; Masson, D. Revisiting the role of LXRs in PUFA metabolism and phospholipid homeostasis. Int. J. Mol. Sci. 2019, 20, 3787.
- Repa, J.J.; Liang, G.; Ou, J.; Bashmakov, Y.; Lobaccaro, J.-M.A.; Shimomura, I.; Shan, B.; Brown, M.S.; Goldstein, J.L.; Mangelsdorf, D.J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000, 14, 2819–2830.
- Seo, J.B.; Moon, H.M.; Kim, W.S.; Lee, Y.S.; Jeong, H.W.; Yoo, E.J.; Ham, J.; Kang, H.; Park, M.-G.; Steffensen, K.R.; et al. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor γ expression. Mol. Cell. Biol. 2004, 24, 3430–3444.
- Cha, J.-Y.; Repa, J.J. The liver X receptor (LXR) and hepatic lipogenesis. J. Biol. Chem. 2007, 282, 743–751.
- Schörken, U.; Kempers, P. Lipid biotechnology: Industrially relevant production processes. Eur. J. Lipid Sci. Technol. 2009, 111, 627–645.
- Shanklin, J.; Cahoon, E.B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 611–641.
- Jayakumar, A.; Tai, M.H.; Huang, W.Y.; Al-Feel, W.; Hsu, M.; Abu-Elheiga, L.; Chirala, S.S.; Wakil, S.J. Human fatty acid synthase: Properties and molecular cloning. Proc. Natl. Acad. Sci. USA 1995, 92, 8695–8699.
- Semenkovich, C.F.; Coleman, T.; Fiedorek, F.T. Human fatty acid synthase mRNA: Tissue distribution, genetic mapping, and kinetics of decay after glucose deprivation. J. Lipid Res. 1995, 36, 1507–1521.
- Nowinski, S.M.; Van Vranken, J.G.; Dove, K.K.; Rutter, J. Impact of mitochondrial fatty acid synthesis on mitochondrial biogenesis. Curr. Biol. 2018, 28, R1212–R1219.
- Strawford, A.; Antelo, F.; Christiansen, M.; Hellerstein, M.K. Adipose tissue triglyceride turnover, de novo lipogenesis, and cell proliferation in humans measured with 2H2O. Am. J. Physiol. Metab. 2004, 286, E577–E588.
- Turner, S.M.; Murphy, E.J.; Neese, R.A.; Antelo, F.; Thomas, T.; Agarwal, A.; Go, C.; Hellerstein, M.K. Measurement of TG synthesis and turnover in vivo by 2 H 2 O incorporation into the glycerol moiety and application of MIDA. Am. J. Physiol. Metab. 2003, 285, E790–E803.
- Bauman, D.E.; Mellenberger, R.W.; Derrig, R.G. Fatty acid synthesis in sheep mammary tissue. J. Dairy Sci. 1973, 56, 1312–1318.
- Chandel, N.S. Glycolysis. Cold Spring Harb. Perspect. Biol. 2021, 13, a040535.
- McCommis, K.S.; Finck, B.N. Mitochondrial pyruvate transport: A historical perspective and future research directions. Biochem. J. 2015, 466, 443–454.
- Martínez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102.
- Wallace, M.; Green, C.R.; Roberts, L.S.; Lee, Y.M.; McCarville, J.L.; Sanchez-Gurmaches, J.; Meurs, N.; Gengatharan, J.M.; Hover, J.D.; Phillips, S.A.; et al. Enzyme promiscuity drives branched-chain fatty acid synthesis in adipose tissues. Nat. Chem. Biol. 2018, 14, 1021–1031.
- Bressler, R.; Wakil, S.J. Studies on the mechanism of fatty acid synthesis. J. Biol. Chem. 1962, 237, 1441–1448.
- Kuhajda, F.P. Fatty-acid synthase and human cancer: New perspectives on its role in tumor biology. Nutrition 2000, 16, 202–208.
- Wakil, S.J. A malonic acid derivative as an intermediate in fatty acid synthesis. J. Am. Chem. Soc. 1958, 80, 6465.
- Wakil, S.J.; Titchener, E.B.; Gibson, D.M. Evidence for the participation of biotin in the enzymic synthesis of fatty acids. Biochim. Biophys. Acta 1958, 29, 225–226.
- O’Neill, L.M.; Miyazaki, M.; Bond, L.M.; Lewis, S.A.; Ding, F.; Liu, Z.; Ntambi, J.M. Fatty acid desaturation and elongation in mammals. In Biochemistry of Lipids, Lipoproteins and Membranes; Ridgway, N.D., McLeod, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 201–226.
- Wakil, S.J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry 1989, 28, 4523–4530.
- Cook, H.W. Fatty acid desaturation and chain elongation in eucaryotes. In Biochemistry of Lipids and Membranes; Vance, D.E., Vance, J.E., Eds.; Elsevier Science: Amsterdam, The Netherlands, 1985; pp. 181–212.
- Heil, C.S.; Wehrheim, S.S.; Paithankar, K.S.; Grininger, M. Fatty acid biosynthesis: Chain-length regulation and control. ChemBioChem 2019, 20, 2298–2321.
- Kim, K.-H. Regulation of mammalian acetyl-coenzyme A carboxylase. Annu. Rev. Nutr. 1997, 17, 77–99.
- Wang, Y.; Yu, W.; Li, S.; Guo, D.; He, J.; Wang, Y. Acetyl-CoA carboxylases and diseases. Front. Oncol. 2022, 12, 836058.
- Paiva, P.; Medina, F.E.; Viegas, M.; Ferreira, P.; Neves, R.P.P.; Sousa, J.P.M.; Ramos, M.J.; Fernandes, P.A. Animal fatty acid synthase: A chemical nanofactory. Chem. Rev. 2021, 121, 9502–9553.
- Leibundgut, M.; Maier, T.; Jenni, S.; Ban, N. The multienzyme architecture of eukaryotic fatty acid synthases. Curr. Opin. Struct. Biol. 2008, 18, 714–725.
- Smith, S. The animal fatty acid synthase: One gene, one polypeptide, seven enzymes. FASEB J. 1994, 8, 1248–1259.
- Magnuson, K.; Jackowski, S.; Rock, C.O.; Cronan, J.E. Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol. Rev. 1993, 57, 522–542.
- Bazan, H.E.P.; Careaga, M.M.; Sprecher, H.; Bazan, N.G. Chain elongation and desaturation of eicosapentaenoate to docosahexaenoate and phospholipid labeling in the rat retina in vivo. Biochim. Biophys. Acta Lipids Lipid Metab. 1982, 712, 123–128.
- Cinti, D.L.; Cook, L.; Nagi, M.N.; Suneja, S.K. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog. Lipid Res. 1992, 31, 1–51.
- Jump, D.B. Mammalian fatty acid elongases. In Lipidomics: Methods in Molecular Biology; Armstrong, D., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 375–389.
- Oboh, A. Investigating the Long-Chain Polyunsaturated Fatty Acid Biosynthesis of the African Catfish Clarias gariepinus (Burchell, 1822). Ph.D. Thesis, University of Stirling, Stirling, UK, 2018.
- Yeboah, G.K.; Lobanova, E.S.; Brush, R.S.; Agbaga, M.-P. Very long chain fatty acid-containing lipids: A decade of novel insights from the study of ELOVL4. J. Lipid Res. 2021, 62, 100030.
- Robinson, B.S.; Johnson, D.W.; Poulos, A. Unique molecular species of phosphatidylcholine containing very-long-chain (C24-C38) polyenoic fatty acids in rat brain. Biochem. J. 1990, 265, 763–767.
- Řezanka, T. Very-long-chain fatty acids from the animal and plant kingdoms. Prog. Lipid Res. 1989, 28, 147–187.
- Poulos, A.; Sharp, P.; Johnson, D.; Easton, C. The occurrence of polyenoic very long chain fatty acids with greater than 32 carbon atoms in molecular species of phosphatidylcholine in normal and peroxisome-deficient (Zellweger’s syndrome) brain. Biochem. J. 1988, 253, 645–650.
- Aveldaño, M.I.; Sprecher, H. Very long chain (C24 to C36) polyenoic fatty acids of the n-3 and n-6 series in dipolyunsaturated phosphatidylcholines from bovine retina. J. Biol. Chem. 1987, 262, 1180–1186.
- Furland, N.E.; Maldonado, E.N.; Aveldaño, M.I. Very long chain PUFA in murine testicular triglycerides and cholesterol esters. Lipids 2003, 38, 73–80.
- Aveldaño, M.I.; Robinson, B.S.; Johnson, D.W.; Poulos, A. Long and very long chain polyunsaturated fatty acids of the n-6 series in rat seminiferous tubules. Active desaturation of 24:4n-6 to 24:5n-6 and concomitant formation of odd and even chain tetraenoic and pentaenoic fatty acids up to C32. J. Biol. Chem. 1993, 268, 11663–11669.
- Furland, N.E.; Zanetti, S.R.; Oresti, G.M.; Maldonado, E.N.; Aveldaño, M.I. Ceramides and sphingomyelins with high proportions of very long-chain polyunsaturated fatty acids in mammalian germ cells. J. Biol. Chem. 2007, 282, 18141–18150.
- Torrissen, M.; Ytteborg, E.; Svensen, H.; Stoknes, I.; Nilsson, A.; Østbye, T.-K.; Berge, G.M.; Bou, M.; Ruyter, B. Investigation of the functions of n-3 very-long-chain PUFAs in skin using in vivo Atlantic salmon and in vitro human and fish skin models. Br. J. Nutr. 2023, 1–17.
- Butovich, I.A. Cholesteryl esters as a depot for very long chain fatty acids in human meibum. J. Lipid Res. 2009, 50, 501–513.
- Butovich, I.A.; Uchiyama, E.; McCulley, J.P. Lipids of human meibum: Mass-spectrometric analysis and structural elucidation. J. Lipid Res. 2007, 48, 2220–2235.
- Rissmann, R.; Groenink, H.W.W.; Weerheim, A.M.; Hoath, S.B.; Ponec, M.; Bouwstra, J.A. New insights into ultrastructure, lipid composition and organization of Vernix Caseosa. J. Investig. Dermatol. 2006, 126, 1823–1833.
- Hiltunen, J.K.; Schonauer, M.S.; Autio, K.J.; Mittelmeier, T.M.; Kastaniotis, A.J.; Dieckmann, C.L. Mitochondrial fatty acid synthesis type II: More than just fatty acids. J. Biol. Chem. 2009, 284, 9011–9015.
- Chuman, L.; Brody, S. Acyl carrier protein is present in the mitochondria of plants and eucaryotic micro-organisms. Eur. J. Biochem. 1989, 184, 643–649.
- Monteuuis, G.; Suomi, F.; Kerätär, J.M.; Masud, A.J.; Kastaniotis, A.J. A conserved mammalian mitochondrial isoform of acetyl-CoA carboxylase ACC1 provides the malonyl-CoA essential for mitochondrial biogenesis in tandem with ACSF3. Biochem. J. 2017, 474, 3783–3797.
- Wongkittichote, P.; Ah Mew, N.; Chapman, K.A. Propionyl-CoA carboxylase—A review. Mol. Genet. Metab. 2017, 122, 145–152.
- Seubert, W.; Podack, E.R. Mechanisms and physiological roles of fatty acid chain elongation in microsomes and mitochondria. Mol. Cell. Biochem. 1973, 1, 29–40.
- Lu, Y.-J.; Zhang, Y.-M.; Grimes, K.D.; Qi, J.; Lee, R.E.; Rock, C.O. Acyl-phosphates initiate membrane phospholipid synthesis in gram-positive pathogens. Mol. Cell 2006, 23, 765–772.
- Rezaei Zonooz, S.; Hasani, M.; Morvaridzadeh, M.; Beatriz Pizarro, A.; Heydari, H.; Yosaee, S.; Rezamand, G.; Heshmati, J. Effect of alpha-lipoic acid on oxidative stress parameters: A systematic review and meta-analysis. J. Funct. Foods 2021, 87, 104774.
- Petersen Shay, K.; Moreau, R.F.; Smith, E.J.; Hagen, T.M. Is α-lipoic acid a scavenger of reactive oxygen speciesin vivo? Evidence for its initiation of stress signaling pathways that promote endogenous antioxidant capacity. IUBMB Life 2008, 60, 362–367.
- Stoll, S.; Hartmann, H.; Cohen, S.A.; Müller, W.E. The potent free radical scavenger α-lipoic acid improves memory in aged mice: Putative relationship to NMDA receptor deficits. Pharmacol. Biochem. Behav. 1993, 46, 799–805.
- Capece, U.; Moffa, S.; Improta, I.; Di Giuseppe, G.; Nista, E.C.; Cefalo, C.M.A.; Cinti, F.; Pontecorvi, A.; Gasbarrini, A.; Giaccari, A.; et al. Alpha-lipoic acid and glucose metabolism: A comprehensive update on biochemical and therapeutic features. Nutrients 2022, 15, 18.
- Hiltunen, J.K.; Autio, K.J.; Schonauer, M.S.; Kursu, V.A.S.; Dieckmann, C.L.; Kastaniotis, A.J. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta Bioenerg. 2010, 1797, 1195–1202.
- Angerer, H.; Schönborn, S.; Gorka, J.; Bahr, U.; Karas, M.; Wittig, I.; Heidler, J.; Hoffmann, J.; Morgner, N.; Zickermann, V. Acyl modification and binding of mitochondrial ACP to multiprotein complexes. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 1913–1920.
- Das, A.K.; Uhler, M.D.; Hajra, A.K. Molecular cloning and expression of mammalian peroxisomaltrans-2-enoyl-coenzyme A reductase cDNAs. J. Biol. Chem. 2000, 275, 24333–24340.
- Buist, P.H. Fatty acid desaturases: Selecting the dehydrogenation channel. Nat. Prod. Rep. 2004, 21, 249.
- Smith, S.; Tsai, S.-C. The type I fatty acid and polyketide synthases: A tale of two megasynthases. Nat. Prod. Rep. 2007, 24, 1041.
- Kaulmann, U.; Hertweck, C. Biosynthesis of polyunsaturated fatty acids by polyketide synthases. Angew. Chem. Int. Ed. 2002, 41, 1866.
- Napier, J. Plumbing the depths of PUFA biosynthesis: A novel polyketide synthase-like pathway from marine organisms. Trends Plant Sci. 2002, 7, 51–54.
- Metz, J.G.; Roessler, P.; Facciotti, D.; Levering, C.; Dittrich, F.; Lassner, M.; Valentine, R.; Lardizabal, K.; Domergue, F.; Yamada, A.; et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 2001, 293, 290–293.
- Chen, Z.; Chen, H.; Li, X.; Yuan, Q.; Su, J.; Yang, L.; Ning, L.; Lei, H. Fumonisin B1 damages the barrier functions of porcine intestinal epithelial cells in vitro. J. Biochem. Mol. Toxicol. 2019, 33, e22397.
- Bentley, R.; Bennett, J.W. Constructing polyketides: From collie to combinatorial biosynthesis. Annu. Rev. Microbiol. 1999, 53, 411–446.
- Cerone, M.; Smith, T.K. Desaturases: Structural and mechanistic insights into the biosynthesis of unsaturated fatty acids. IUBMB Life 2022, 74, 1036–1051.
- Gostinčar, C.; Turk, M.; Gunde-Cimerman, N. The evolution of fatty acid desaturases and cytochrome b5 in eukaryotes. J. Membr. Biol. 2010, 233, 63–72.
- Sprecher, H.; Luthria, D.L.; Mohammed, B.S.; Baykousheva, S.P. Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. J. Lipid Res. 1995, 36, 2471–2477.
- Paton, C.M.; Ntambi, J.M. Biochemical and physiological function of stearoyl-CoA desaturase. Am. J. Physiol. Metab. 2009, 297, E28–E37.
- Nakamura, M.T.; Nara, T.Y. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu. Rev. Nutr. 2004, 24, 345–376.
- Meesapyodsuk, D.; Qiu, X. Structure Determinants for the Substrate Specificity of Acyl-CoA Δ9 Desaturases from a Marine Copepod. ACS Chem. Biol. 2014, 9, 922–934.
- Haritos, V.S.; Horne, I.; Damcevski, K.; Glover, K.; Gibb, N. Unexpected functional diversity in the fatty acid desaturases of the flour beetle Tribolium castaneum and identification of key residues determining activity. Insect Biochem. Mol. Biol. 2014, 51, 62–70.
- Bonamore, A.; Macone, A.; Colotti, G.; Matarese, R.M.; Boffi, A. The desaturase from Bacillus subtilis, a promising tool for the selective olefination of phospholipids. J. Biotechnol. 2006, 121, 49–53.
- Heilmann, I.; Mekhedov, S.; King, B.; Browse, J.; Shanklin, J. Identification of the Arabidopsis Palmitoyl-Monogalactosyldiacylglycerol Δ7-Desaturase Gene FAD5, and Effects of Plastidial Retargeting of Arabidopsis Desaturases on the fad5 Mutant Phenotype. Plant Physiol. 2004, 136, 4237–4245.
- Knipple, D.C.; Rosenfield, C.-L.; Miller, S.J.; Liu, W.; Tang, J.; Ma, P.W.K.; Roelofs, W.L. Cloning and functional expression of a cDNA encoding a pheromone gland-specific acyl-CoA Δ11-desaturase of the cabbage looper moth, Trichoplusia ni. Proc. Natl. Acad. Sci. USA 1998, 95, 15287–15292.
- Ohnishi, M.; Thompson, G.A. Biosynthesis of the unique trans-Δ3-hexadecenoic acid component of chloroplast phosphatidylglycerol: Evidence concerning its site and mechanism of formation. Arch. Biochem. Biophys. 1991, 288, 591–599.
- Weiss-Hersh, K.; Garcia, A.L.; Marosvölgyi, T.; Szklenár, M.; Decsi, T.; Rühl, R. Saturated and monounsaturated fatty acids in membranes are determined by the gene expression of their metabolizing enzymes SCD1 and ELOVL6 regulated by the intake of dietary fat. Eur. J. Nutr. 2020, 59, 2759–2769.
- Miyazaki, M.; Jacobson, M.J.; Man, W.C.; Cohen, P.; Asilmaz, E.; Friedman, J.M.; Ntambi, J.M. Identification and characterization of murine SCD4, a novel heart-specific stearoyl-CoA desaturase isoform regulated by leptin and dietary factors. J. Biol. Chem. 2003, 278, 33904–33911.
- Human Gene Nomenclature Containing (HGNC) Gene Group: Fatty Acid Desaturases (FADS). Available online: https://www.genenames.org/data/genegroup/#!/group/553 (accessed on 15 September 2023).
- Nagao, K.; Murakami, A.; Umeda, M. Structure and Function of Δ9-Fatty Acid Desaturase. Chem. Pharm. Bull. 2019, 67, 327–332.
- Grajchen, E.; Loix, M.; Baeten, P.; Côrte-Real, B.F.; Hamad, I.; Vanherle, S.; Haidar, M.; Dehairs, J.; Broos, J.Y.; Ntambi, J.M.; et al. Fatty acid desaturation by stearoyl-CoA desaturase-1 controls regulatory T cell differentiation and autoimmunity. Cell. Mol. Immunol. 2023, 20, 666–679.
- Tang, B.; Qiu, J.; Hu, S.; Li, L.; Wang, J. Role of stearyl-coenzyme A desaturase 1 in mediating the effects of palmitic acid on endoplasmic reticulum stress, inflammation, and apoptosis in goose primary hepatocytes. Anim. Biosci. 2021, 34, 1210–1220.
- Koeberle, A.; Löser, K.; Thürmer, M. Stearoyl-CoA desaturase-1 and adaptive stress signaling. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2016, 1861, 1719–1726.
- Kucharski, M.; Kaczor, U. Stearoyl-CoA desaturase—The lipid metabolism regulator. Adv. Hyg. Exp. Med. 2014, 68, 334–342.
- Liu, X.; Strable, M.S.; Ntambi, J.M. Stearoyl CoA Desaturase 1: Role in Cellular Inflammation and Stress. Adv. Nutr. 2011, 2, 15–22.
- Dobrzyn, P.; Dobrzyn, A.; Miyazaki, M.; Cohen, P.; Asilmaz, E.; Hardie, D.G.; Friedman, J.M.; Ntambi, J.M. Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proc. Natl. Acad. Sci. USA 2004, 101, 6409–6414.
- Park, W.J.; Kothapalli, K.S.D.; Reardon, H.T.; Lawrence, P.; Qian, S.-B.; Brenna, J.T. A novel FADS1 isoform potentiates FADS2-mediated production of eicosanoid precursor fatty acids. J. Lipid Res. 2012, 53, 1502–1512.
- Park, W.J.; Reardon, H.T.; Tyburczy, C.; Kothapalli, K.S.D.; Brenna, J.T. Alternative splicing generates a novel FADS2 alternative transcript in baboons. Mol. Biol. Rep. 2010, 37, 2403–2406.
- Park, W.J.; Kothapalli, K.S.D.; Reardon, H.T.; Kim, L.Y.; Brenna, J.T. Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene 2009, 446, 28–34.
- Srikanth, K.; Kwan, A.; Lee, E.; Kim, S.; Lim, Y.; Chung, H. Associations of single nucleotide polymorphisms in the bovine FADS6 gene with fatty acid composition in Hanwoo (Korean Cattle). Open J. Genet. 2015, 05, 137–144.
- Chen, H.; Hao, G.; Wang, L.; Wang, H.; Gu, Z.; Liu, L.; Zhang, H.; Chen, W.; Chen, Y.Q. Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci. Rep. 2015, 5, 11247.
- Stroud, C.K.; Nara, T.Y.; Roqueta-Rivera, M.; Radlowski, E.C.; Lawrence, P.; Zhang, Y.; Cho, B.H.; Segre, M.; Hess, R.A.; Brenna, J.T.; et al. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 2009, 50, 1870–1880.
- Zhu, K.-C.; Song, L.; Guo, H.-Y.; Guo, L.; Zhang, N.; Liu, B.-S.; Jiang, S.-G.; Zhang, D.-C. Identification of fatty acid desaturase 6 in Golden Pompano Trachinotus Ovatus (Linnaeus 1758) and its regulation by the PPARαb transcription factor. Int. J. Mol. Sci. 2018, 20, 23.
- Marquardt, A.; Stöhr, H.; White, K.; Weber, B.H.F. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics 2000, 66, 175–183.
- Rioux, V.; Pédrono, F.; Blanchard, H.; Duby, C.; Boulier-Monthéan, N.; Bernard, L.; Beauchamp, E.; Catheline, D.; Legrand, P. Trans-vaccenate is Δ13-desaturated by FADS3 in rodents. J. Lipid Res. 2013, 54, 3438–3452.
- Zhang, J.Y.; Qin, X.; Liang, A.; Kim, E.; Lawrence, P.; Park, W.J.; Kothapalli, K.S.D.; Brenna, J.T. Fads3 modulates docosahexaenoic acid in liver and brain. Prostaglandins Leukot. Essent. Fat. Acids 2017, 123, 25–32.
- Karsai, G.; Lone, M.; Kutalik, Z.; Brenna, J.T.; Li, H.; Pan, D.; von Eckardstein, A.; Hornemann, T. FADS3 is a Δ14Z sphingoid base desaturase that contributes to gender differences in the human plasma sphingolipidome. J. Biol. Chem. 2020, 295, 1889–1897.
- Castro, L.F.C.; Tocher, D.R.; Monroig, O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 2016, 62, 25–40.
- Lee, J.; Lee, H.; Kang, S.; Park, W. Fatty acid desaturases, polyunsaturated fatty acid regulation, and biotechnological advances. Nutrients 2016, 8, 23.
- Guillou, H.; Zadravec, D.; Martin, P.G.P.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199.
- Spector, A.A.; Kim, H.-Y. Discovery of essential fatty acids. J. Lipid Res. 2015, 56, 11–21.
- Kaur, N.; Chugh, V.; Gupta, A.K. Essential fatty acids as functional components of foods—A review. J. Food Sci. Technol. 2014, 51, 2289–2303.
- Burr, G.O.; Burr, M.M. On the nature and role of the fatty acids essential in nutrition. J. Biol. Chem. 1930, 86, 587–621.
- Cho, H.P.; Nakamura, M.; Clarke, S.D. Cloning, expression, and fatty acid regulation of the human Δ-5 desaturase. J. Biol. Chem. 1999, 274, 37335–37339.
- Cho, H.P.; Nakamura, M.T.; Clarke, S.D. Cloning, expression, and nutritional regulation of the mammalian Δ-6 desaturase. J. Biol. Chem. 1999, 274, 471–477.
- Scott, B.L.; Bazan, N.G. Membrane docosahexaenoate is supplied to the developing brain and retina by the liver. Proc. Natl. Acad. Sci. USA 1989, 86, 2903–2907.
- De Antueno, R.J.; Knickle, L.C.; Smith, H.; Elliot, M.L.; Allen, S.J.; Nwaka, S.; Winther, M.D. Activity of human Δ5 and Δ6 desaturases on multiple n-3 and n-6 polyunsaturated fatty acids. FEBS Lett. 2001, 509, 77–80.
- Vagner, M.; Santigosa, E. Characterization and modulation of gene expression and enzymatic activity of delta-6 desaturase in teleosts: A review. Aquaculture 2011, 315, 131–143.
- Arshad, Z.; Rezapour-Firouzi, S.; Ebrahimifar, M.; Mosavi-Jarrahi, A.; Mohammadian, M. Association of delta-6-desaturase expression with aggressiveness of cancer, diabetes mellitus, and multiple sclerosis: A narrative review. Asian Pac. J. Cancer Prev. 2019, 20, 1005–1018.
- Tosi, F.; Sartori, F.; Guarini, P.; Olivieri, O.; Martinelli, N. Delta-5 and delta-6 desaturases: Crucial enzymes in polyunsaturated fatty acid-related pathways with pleiotropic influences in health and disease. In Oxidative Stress and Inflammation in Non-Communicable Diseases—Molecular Mechanisms and Perspectives in Therapeutics: Advances in Experimental Medicine and Biology; Camps, J., Ed.; Springer: Cham, Switzerland, 2014; pp. 61–81.
- Mead, J.F.; Slaton, W.H. Metabolism of essential fatty acids. J. Biol. Chem. 1956, 219, 705–709.
- Siguel, E.N.; Chee, K.M.; Gong, J.X.; Schaefer, E.J. Criteria for essential fatty acid deficiency in plasma as assessed by capillary column gas-liquid chromatography. Clin. Chem. 1987, 33, 1869–1873.
- Holman, R.T. The ratio of trienoic: Tetraenoic acids in tissue lipids as a measure of essential fatty acid requirement. J. Nutr. 1960, 70, 405–410.
- Park, H.G.; Engel, M.G.; Vogt-Lowell, K.; Lawrence, P.; Kothapalli, K.S.; Brenna, J.T. The role of fatty acid desaturase (FADS) genes in oleic acid metabolism: FADS1 Δ7 desaturates 11-20:1 to 7,11-20:2. Prostaglandins Leukot. Essent. Fat. Acids 2018, 128, 21–25.
- Park, H.G.; Park, W.J.; Kothapalli, K.S.D.; Brenna, J.T. The fatty acid desaturase 2 (FADS2) gene product catalyzes Δ4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells. FASEB J. 2015, 29, 3911–3919.
- Li, Y.; Monroig, O.; Zhang, L.; Wang, S.; Zheng, X.; Dick, J.R.; You, C.; Tocher, D.R. Vertebrate fatty acyl desaturase with Δ4 activity. Proc. Natl. Acad. Sci. USA 2010, 107, 16840–16845.
- Martinez, M.; Ichaso, N.; Setien, F.; Durany, N.; Qiu, X.; Roesler, W. The Δ4-desaturation pathway for DHA biosynthesis is operative in the human species: Differences between normal controls and children with the Zellweger syndrome. Lipids Health Dis. 2010, 9, 98.
- Qiu, X.; Hong, H.; MacKenzie, S.L. Identification of a Δ4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by Heterologous Expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 2001, 276, 31561–31566.
- Schenck, P.A.; Rakoff, H.; Emken, E.A. δ8 desaturationin vivo of deuterated eicosatrienoic acid by mouse liver. Lipids 1996, 31, 593–600.
- Cook, H.; Byers, D.; Palmer, F.; Spence, M.; Rakoff, H.; Duval, S.; Emken, E. Alternate pathways in the desaturation and chain elongation of linolenic acid, 18:3(n-3), in cultured glioma cells. J. Lipid Res. 1991, 32, 1265–1273.
- Park, W.J.; Kothapalli, K.S.D.; Lawrence, P.; Tyburczy, C.; Brenna, J.T. An alternate pathway to long-chain polyunsaturates: The FADS2 gene product Δ8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 2009, 50, 1195–1202.
- Kabeya, N.; Fonseca, M.M.; Ferrier, D.E.K.; Navarro, J.C.; Bay, L.K.; Francis, D.S.; Tocher, D.R.; Castro, L.F.C.; Monroig, Ó. Genes for de novo biosynthesis of omega-3 polyunsaturated fatty acids are widespread in animals. Sci. Adv. 2018, 4, eaar6849.
- Xue, Z.; He, H.; Hollerbach, D.; Macool, D.J.; Yadav, N.S.; Zhang, H.; Szostek, B.; Zhu, Q. Identification and characterization of new Δ-17 fatty acid desaturases. Appl. Microbiol. Biotechnol. 2013, 97, 1973–1985.
- Jump, D.B.; Clarke, S.D.; Thelen, A.; Liimatta, M.; Ren, B.; Badin, M. Dietary polyunsaturated fatty acid regulation of gene transcription. Prog. Lipid Res. 1996, 35, 227–241.
- Blake, W.L.; Clarke, S.D. Suppression of rat hepatic fatty acid synthase and S14 gene transcription by dietary polyunsaturated fat. J. Nutr. 1990, 120, 1727–1729.
- Mullen, T.D.; Hannun, Y.A.; Obeid, L.M. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem. J. 2012, 441, 789–802.
- Brush, R.S.; Tran, J.-T.A.; Henry, K.R.; McClellan, M.E.; Elliott, M.H.; Mandal, M.N.A. Retinal sphingolipids and their very-long-chain fatty acid–containing species. Investig. Opthalmology Vis. Sci. 2010, 51, 4422.
- Sandhoff, R. Very long chain sphingolipids: Tissue expression, function and synthesis. FEBS Lett. 2010, 584, 1907–1913.
- Nachtschatt, M.; Okada, S.; Speight, R. Integral membrane fatty acid desaturases: A review of biochemical, structural, and biotechnological advances. Eur. J. Lipid Sci. Technol. 2020, 122, 2000181.
- Zhu, G.; Koszelak-Rosenblum, M.; Connelly, S.M.; Dumont, M.E.; Malkowski, M.G. The crystal structure of an integral membrane fatty acid α-hydroxylase. J. Biol. Chem. 2015, 290, 29820–29833.
- Vacchina, P.; Tripodi, K.E.J.; Escalante, A.M.; Uttaro, A.D. Characterization of bifunctional sphingolipid Δ4-desaturases/C4-hydroxylases of trypanosomatids by liquid chromatography–electrospray tandem mass spectrometry. Mol. Biochem. Parasitol. 2012, 184, 29–38.
- Chen, M.; Markham, J.E.; Cahoon, E.B. Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012, 69, 769–781.
- López-Lara, I.M.; Soto, M.J. Fatty acid synthesis and regulation. In Biogenesis of Fatty Acids, Lipids and Membranes; Springer International Publishing: Cham, Switzerland, 2019; pp. 391–407.
- Yamashita, A.; Hayashi, Y.; Nemoto-Sasaki, Y.; Ito, M.; Oka, S.; Tanikawa, T.; Waku, K.; Sugiura, T. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 2014, 53, 18–81.
- Kent, C. Eukaryotic phospholipid biosynthesis. Annu. Rev. Biochem. 1995, 64, 315–343.
- Quehenberger, O.; Armando, A.M.; Brown, A.H.; Milne, S.B.; Myers, D.S.; Merrill, A.H.; Bandyopadhyay, S.; Jones, K.N.; Kelly, S.; Shaner, R.L.; et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010, 51, 3299–3305.
- Shindou, H.; Hishikawa, D.; Harayama, T.; Yuki, K.; Shimizu, T. Recent progress on acyl CoA: Lysophospholipid acyltransferase research. J. Lipid Res. 2009, 50, S46–S51.
- Coleman, R.A.; Lewin, T.M.; Van Horn, C.G.; Gonzalez-Baró, M.R. Do long-chain acyl-CoA synthetases regulate fatty acid Entry into Synthetic Versus degradative pathways? J. Nutr. 2002, 132, 2123–2126.
- MacDonald, J.I.S.; Sprecher, H. Phospholipid fatty acid remodeling in mammalian cells. Biochim. Biophys. Acta Lipids Lipid Metab. 1991, 1084, 105–121.
- Vance, J.E. Historical perspective: Phosphatidylserine and phosphatidylethanolamine from the 1800s to the present. J. Lipid Res. 2018, 59, 923–944.
- Lands, W.E.M. Stories about acyl chains. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2000, 1483, 1–14.
- Lands, W.E.M.; Merkl, I. Metabolism of glycerolipids. J. Biol. Chem. 1963, 238, 898–904.
- Markham, J.E.; Molino, D.; Gissot, L.; Bellec, Y.; Hématy, K.; Marion, J.; Belcram, K.; Palauqui, J.-C.; Satiat-JeuneMaître, B.; Faure, J.-D. Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in arabidopsis. Plant Cell 2011, 23, 2362–2378.
- Samovski, D.; Jacome-Sosa, M.; Abumrad, N.A. Fatty Acid transport and signaling: Mechanisms and physiological implications. Annu. Rev. Physiol. 2023, 85, 317–337.
- Glatz, J.F.C.; Luiken, J.J.F.P.; Bonen, A. Membrane fatty acid transporters as regulators of lipid metabolism: Implications for metabolic disease. Physiol. Rev. 2010, 90, 367–417.
- Farkas, T. Adaptation of fatty acid compositions to temperature—A study on planktonic crustaceans. Comp. Biochem. Physiol. Part B Comp. Biochem. 1979, 64, 71–76.
- Dymond, M.K. Mammalian phospholipid homeostasis: Evidence that membrane curvature elastic stress drives homeoviscous adaptation in vivo. J. R. Soc. Interface 2016, 13, 20160228.
- Kulig, W.; Pasenkiewicz-Gierula, M.; Róg, T. Cis and trans unsaturated phosphatidylcholine bilayers: A molecular dynamics simulation study. Chem. Phys. Lipids 2016, 195, 12–20.
- Gillan, F.T.; Johns, R.B.; Verheyen, T.V.; Volkman, J.K.; Bavor, H.J. Trans-monounsaturated acids in a marine bacterial isolate. Appl. Environ. Microbiol. 1981, 41, 849–856.
- Hanahan, D.J.; Brockerhoff, H.; Barron, E.J. The Site of Attack of Phospholipase (Lecithinase) A on Lecithin: A Re-evaluation. J. Biol. Chem. 1960, 235, 1917–1923.
- Tattrie, N.H. Positional distribution of saturated and unsaturated fatty acids on egg lecithin. J. Lipid Res. 1959, 1, 60–65.
- Pearson, R.H.; Pascher, I. The molecular structure of lecithin dihydrate. Nature 1979, 281, 499–501.
- Büldt, G.; Gally, H.U.; Seelig, J.; Zaccai, G. Neutron diffraction studies on phosphatidylcholine model membranes. J. Mol. Biol. 1979, 134, 673–691.
- Sanders, R.L.; Longmore, W.J. Phosphatidylglycerol in rat lung. II. Comparison of occurrence, composition, and metabolism in surfactant and residual lung fractions. Biochemistry 1975, 14, 835–840.
- Xie, D.; Seremwe, M.; Edwards, J.G.; Podolsky, R.; Bollag, W.B. Distinct Effects of Different Phosphatidylglycerol Species on Mouse Keratinocyte Proliferation. PLoS ONE 2014, 9, e107119.
- O’Donnell, V.B. New appreciation for an old pathway: The Lands Cycle moves into new arenas in health and disease. Biochem. Soc. Trans. 2022, 50, 1–11.
- Nakanishi, H.; Iida, Y.; Shimizu, T.; Taguchi, R. Separation and quantification of sn-1 and sn-2 fatty acid positional isomers in phosphatidylcholine by RPLC-ESIMS/MS. J. Biochem. 2010, 147, 245–256.
- Wood, R.; Harlow, R.D. Structural studies of neutral glycerides and phosphoglycerides of rat liver. Arch. Biochem. Biophys. 1969, 131, 495–501.
- Kuksis, A.; Breckenridge, W.C.; Marai, L.; Stachnyk, O. Molecular species of lecithins of rat heart, kidney, and plasma. J. Lipid Res. 1969, 10, 25–32.
- Yabuuchi, H.; O’Brien, J.S. Positional distribution of fatty acids in glycerophosphatides of bovine gray matter. J. Lipid Res. 1968, 9, 65–67.
- O’Brien, J.S.; Rouser, G. The fatty acid composition of brain sphingolipids: Sphingomyelin, ceramide, cerebroside, and cerebroside sulfate. J. Lipid Res. 1964, 5, 339–342.
- Kuksis, A.; Marai, L. Determination of the complete structure of natural lecithins. Lipids 1967, 2, 217–224.
- Holub, B.J.; Kuksis, A. Molecular species of phosphatidyl ethanolamine from egg yolk. Lipids 1969, 4, 466–472.
- Doğru Pekiner, B. Fatty acid composition of red blood cell membrane phosphatidylethanolamine and phosphatidylcholine in the rat, rabbit, human and dog: Sıçan, tavşan, insan ve köpek eritrosit membranı fosfatidiletanol. Ankara Univ. Eczac. Fak. Derg. 2002, 31, 169–182.
- Kim, H.-Y.; Huang, B.X.; Spector, A.A. Phosphatidylserine in the brain: Metabolism and function. Prog. Lipid Res. 2014, 56, 1–18.
- Hamilton, J.; Greiner, R.; Salem, N.; Kim, H.-Y. n-3 Fatty acid deficiency decreases phosphatidylserine accumulation selectively in neuronal tissues. Lipids 2000, 35, 863–869.
- Skotland, T.; Sandvig, K. The role of PS 18:0/18:1 in membrane function. Nat. Commun. 2019, 10, 2752.
- Clark, S.R.; Thomas, C.P.; Hammond, V.J.; Aldrovandi, M.; Wilkinson, G.W.; Hart, K.W.; Murphy, R.C.; Collins, P.W.; O’Donnell, V.B. Characterization of platelet aminophospholipid externalization reveals fatty acids as molecular determinants that regulate coagulation. Proc. Natl. Acad. Sci. USA 2013, 110, 5875–5880.
- Thompson, W.; MacDonald, G. Isolation and characterization of cytidine diphosphate diglyceride from beef liver. J. Biol. Chem. 1975, 250, 6779–6785.
- Holub, B.J.; Kuksis, A.; Thompson, W. Molecular species of mono-, di-, and triphosphoinositides of bovine brain. J. Lipid Res. 1970, 11, 558–564.
- Ulmann, L.; Mimouni, V.; Roux, S.; Porsolt, R.; Poisson, J.-P. Brain and hippocampus fatty acid composition in phospholipid classes of aged-relative cognitive deficit rats. Prostaglandins Leukot. Essent. Fat. Acids 2001, 64, 189–195.
- Gijón, M.A.; Riekhof, W.R.; Zarini, S.; Murphy, R.C.; Voelker, D.R. Lysophospholipid Acyltransferases and Arachidonate Recycling in Human Neutrophils. J. Biol. Chem. 2008, 283, 30235–30245.
- Ridgway, N.D. Phospholipid synthesis in mammalian cells. In Biochemistry of Lipids, Lipoproteins and Membranes; Ridgway, N.D., McLeod, R.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 227–258.
- Jia, D.; Zhang, J.; Nie, J.; Andersen, J.-P.; Rendon, S.; Zheng, Y.; Liu, X.; Tian, Z.; Shi, Y. Cardiolipin remodeling by ALCAT1 links hypoxia to coronary artery disease by promoting mitochondrial dysfunction. Mol. Ther. 2021, 29, 3498–3511.
- Pennington, E.R.; Funai, K.; Brown, D.A.; Shaikh, S.R. The role of cardiolipin concentration and acyl chain composition on mitochondrial inner membrane molecular organization and function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1039–1052.
- Li, J.; Romestaing, C.; Han, X.; Li, Y.; Hao, X.; Wu, Y.; Sun, C.; Liu, X.; Jefferson, L.S.; Xiong, J.; et al. Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell Metab. 2010, 12, 154–165.
- Stefanyk, L.E.; Coverdale, N.; Roy, B.D.; Peters, S.J.; LeBlanc, P.J. Skeletal muscle type comparison of subsarcolemmal mitochondrial membrane phospholipid fatty acid composition in rat. J. Membr. Biol. 2010, 234, 207–215.
- Schlame, M.; Ren, M.; Xu, Y.; Greenberg, M.L.; Haller, I. Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids 2005, 138, 38–49.
- Berger, A.; Gershwin, M.E.; German, J.B. Effects of various dietary fats on cardiolipin acyl composition during ontogeny of mice. Lipids 1992, 27, 605–612.
- Wolff, R.L.; Entressangles, B. Compositional changes of fatty acids in the 1(1″)- and 2(2″)-positions of cardiolipin from liver, heart, and kidney mitochondria of rats fed a low-fat diet. Biochim. Biophys. Acta Lipids Lipid Metab. 1991, 1082, 136–142.
- Brotherus, J.; Renkonen, O. Isolation and characterisation of bis-phosphatidic acid and its partially deacylated derivatives from cultured BHK-cells. Chem. Phys. Lipids 1974, 13, 11–20.
- Akgoc, Z.; Iosim, S.; Seyfried, T.N. Bis(monoacylglycero)phosphate as a macrophage enriched phospholipid. Lipids 2015, 50, 907–912.
- Anderson, D.M.G.; Ablonczy, Z.; Koutalos, Y.; Hanneken, A.M.; Spraggins, J.M.; Calcutt, M.W.; Crouch, R.K.; Caprioli, R.M.; Schey, K.L. Bis(monoacylglycero)phosphate lipids in the retinal pigment epithelium implicate lysosomal/endosomal dysfunction in a model of Stargardt disease and human retinas. Sci. Rep. 2017, 7, 17352.
- Besson, N.; Hullin-Matsuda, F.; Makino, A.; Murate, M.; Lagarde, M.; Pageaux, J.-F.; Kobayashi, T.; Delton-Vandenbroucke, I. Selective incorporation of docosahexaenoic acid into lysobisphosphatidic acid in cultured THP-1 macrophages. Lipids 2006, 41, 189–196.
- Luquain, C.; Dolmazon, R.; Enderlin, J.M.; Laugier, C.; Lagarde, M.; Pageaux, J.F. Bis(monoacylglycerol) phosphate in rat uterine stromal cells: Structural characterization and specific esterification of docosahexaenoic acid. Biochem. J. 2000, 351 Pt 3, 795–804.
- Huterer, S.; Wherrett, J. Metabolism of bis(monoacylglycero)phosphate in macrophages. J. Lipid Res. 1979, 20, 966–973.
- Wherrett, J.R.; Huterer, S. Bis-(monoacylglyceryl)-phosphate of rat and human liver: Fatty acid composition and NMR spectroscopy. Lipids 1973, 8, 531–533.
- Holbrook, P.G.; Pannell, L.K.; Murata, Y.; Daly, J.W. Bis(monoacylglycero) phosphate from PC12 cells, a phospholipid that can comigrate with phosphatidic acid: Molecular species analysis by fast atom bombardment mass spectrometry. Biochim. Biophys. Acta Lipids Lipid Metab. 1992, 1125, 330–334.
- Ryan, S.D.; Harris, C.S.; Carswell, C.L.; Baenziger, J.E.; Bennett, S.A.L. Heterogeneity in the sn-1 carbon chain of platelet-activating factor glycerophospholipids determines pro- or anti-apoptotic signaling in primary neurons. J. Lipid Res. 2008, 49, 2250–2258.
- Nakagawa, Y.; Horrocks, L.A. Different metabolic rates for arachidonoyl molecular species of ethanolamine glycerophospholipids in rat brain. J. Lipid Res. 1988, 27, 629–636.
- Tomonaga, N.; Tsuduki, T.; Manabe, Y.; Sugawara, T. Sphingoid bases of dietary ceramide 2-aminoethylphosphonate, a marine sphingolipid, absorb into lymph in rats. J. Lipid Res. 2019, 60, 333–340.
- Pruett, S.T.; Bushnev, A.; Hagedorn, K.; Adiga, M.; Haynes, C.A.; Sullards, M.C.; Liotta, D.C.; Merrill, A.H. Thematic review series: Sphingolipids. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J. Lipid Res. 2008, 49, 1621–1639.
- Merrill, A.H.; Nixon, D.W.; Williams, R.D. Activities of serine palmitoyltransferase (3-ketosphinganine synthase) in microsomes from different rat tissues. J. Lipid Res. 1985, 26, 617–622.
- Ardail, D.; Popa, I.; Alcantara, K.; Pons, A.; Zanetta, J.; Louisot, P.; Thomas, L.; Portoukalian, J. Occurrence of ceramides and neutral glycolipids with unusual long-chain base composition in purified rat liver mitochondria. FEBS Lett. 2001, 488, 160–164.
- Lingwood, D.; Simons, K. Lipid rafts as a membrane-organizing principle. Science 2010, 327, 46–50.
- Eckhardt, M. Fatty Acid 2-Hydroxylase and 2-Hydroxylated Sphingolipids: Metabolism and Function in Health and Diseases. Int. J. Mol. Sci. 2023, 24, 4908.
- Marquês, J.T.; Marinho, H.S.; de Almeida, R.F.M. Sphingolipid hydroxylation in mammals, yeast and plants—An integrated view. Prog. Lipid Res. 2018, 71, 18–42.
- Sessa, L.; Nardiello, A.M.; Santoro, J.; Concilio, S.; Piotto, S. Hydroxylated fatty acids: The role of the sphingomyelin synthase and the origin of selectivity. Membranes 2021, 11, 787.
- Barenholz, Y. Sphingomyelin-lecithin balance in membranes: Composition, structure, and function relationships. In Physiology of Membrane Fluidity; Shinitzky, M., Ed.; CRC Press: Boca Raton, FL, USA, 1984; Volume 1, pp. 131–173.
- Ramstedt, B.; Leppimäki, P.; Axberg, M.; Slotte, J.P. Analysis of natural and synthetic sphingomyelins using high-performance thin-layer chromatography. Eur. J. Biochem. 1999, 266, 997–1002.
- Karlsson, A.Å.; Michélsen, P.; Odham, G. Molecular species of sphingomyelin: Determination by high-performance liquid chromatography/mass spectrometry with electrospray and high-performance liquid chromatography/tandem mass spectrometry with atmospheric pressure chemical ionization. J. Mass Spectrom. 1998, 33, 1192–1198.
- Calhoun, W.I.; Shipley, G.G. Fatty acid composition and thermal behavior of natural sphingomyelins. Biochim. Biophys. Acta Biomembr. 1979, 555, 436–441.
- Poulos, A.; Sharp, P.; Johnson, D.; White, I.; Fellenberg, A. The occurrence of polyenoic fatty acids with greater than 22 carbon atoms in mammalian spermatozoa. Biochem. J. 1986, 240, 891–895.
- Robinson, B.S.; Johnson, D.W.; Poulos, A. Novel molecular species of sphingomyelin containing 2-hydroxylated polyenoic very-long-chain fatty acids in mammalian testes and spermatozoa. J. Biol. Chem. 1992, 267, 1746–1751.
- Skotland, T.; Sandvig, K. Need for more focus on lipid species in studies of biological and model membranes. Prog. Lipid Res. 2022, 86, 101160.
- Fujiwara, Y.; Hama, K.; Yokoyama, K. Mass spectrometry in combination with a chiral column and multichannel-MRM allows comprehensive analysis of glycosphingolipid molecular species from mouse brain. Carbohydr. Res. 2020, 490, 107959.
- Baumann, N.; Pham-Dinh, D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001, 81, 871–927.
- Klenk, E.; Hendricks, U.W.; Gielen, W. β-D-Galaktosido-(1->3)-N-acetyl-D-galaktosamin, ein kristallisiertes Disaccharid aus menschlichen Gehirngangliosiden. Biol. Chem. 1962, 330, 140–144.
- Sastry, P.S. Lipids of nervous tissue: Composition and metabolism. Prog. Lipid Res. 1985, 24, 69–176.
