Alzheimer’s disease (AD) is the most common neurodegenerative disorder globally. In people aged 65 and older, it is estimated that 1 in 9 currently live with the disease. With aging being the greatest risk factor for disease onset, the physiological, social and economic burden continues to rise. Thus, AD remains a public health priority. Since 2007, genome-wide association studies (GWAS) have identified over 80 genomic loci with variants associated with increased AD risk. Although some variants are beginning to be characterized, the effects of many risk loci remain to be elucidated. One advancement which may help provide a patient-focused approach to tackle this issue is the application of gene editing technology and human-induced pluripotent stem cells (hiPSCs).
- Alzheimer’s disease
- GWAS
- iPSC
- microglia
- neurodegeneration
1. Introduction to Alzheimer’s Disease
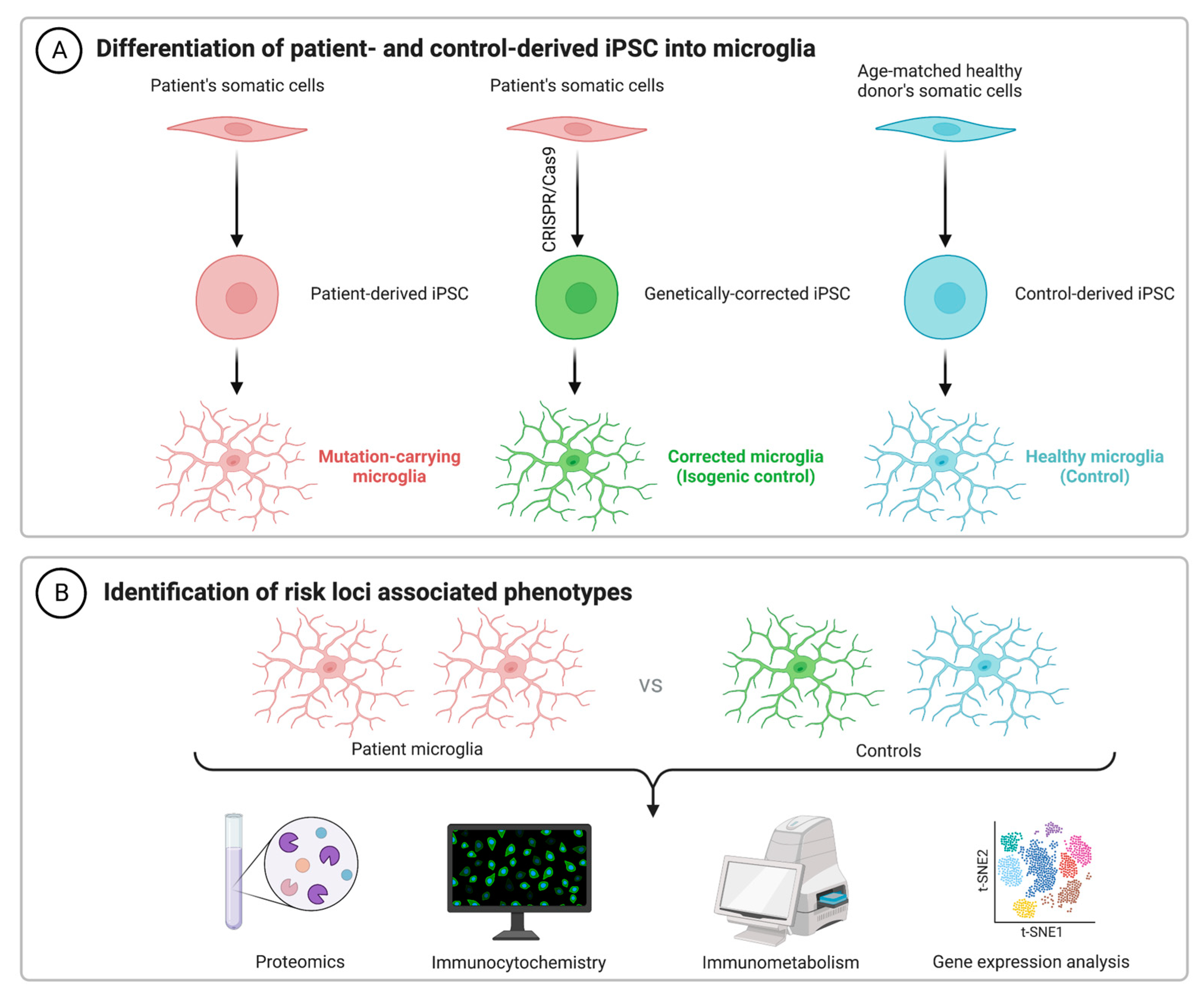
2. iPSC Technology and iPSC-Derived Microglia
As an alternative to using immortalized lines, primary cells from animal models and human microglia from brain excision surgery, recent advances in technology have facilitated a renewed interested in the generation of microglia from human-induced pluripotent stem cells (hiPSCs). Getting around the issue of tissue availability and variation in microglial phenotypes, hiPSCs can be used for both in vitro and xenographic in vivo work to understand cellular physiology in both health and disease. For example, microglial chimeric mouse models have been successfully generated, wherein hiPSC-derived macrophage progenitors are transplanted into neonatal mouse brains. Single-cell RNA sequencing reported that these xenografted cells largely retain human microglial identity and become widely dispersed by 6 months of age, including in both the hippocampus and cerebral cortex, as reported by TMEM119 staining [82][20]. It has been suggested that TMEM119 is both a reliable and specific marker, able to discriminate resident microglia from peripherally derived macrophages [83,84][21][22]. However, recent work suggests that TMEM119 is not exclusive, nor does it stain all microglia. The latter appears especially true under cellular stress conditions, including murine models of both focal stroke and Parkinson’s disease [85][23]. As such, supplementary staining methods will be important for future work using this model. Despite this issue though, in tandem with murine microglial depletion methods such as the pharmacological inhibition of CSF1R [86[24][25],87], the function of humanized microglia can be validated and compared against traditional non-chimeric animals. Pharmacological and genetic studies which report benefits in both systems will likely strengthen the translational potential of any promising future therapeutic target. Although the understanding of microglial function continues to improve, cellular dysfunction in AD remains to be fully elucidated, despite the identification of dozens of genetic risk loci (Table 1). The use of hiPSCs to decipher the pathological contributions of these risk variants could thus prove invaluable. Following the seminal publication by Muffet and colleagues in 2016 [88][26], many protocols to produce hiPSC-derived microglia have become available [89,90,91,92[27][28][29][30][31][32],93,94], with others having reviewed some of these protocols previously [95][33].| Gene | SNPs | GWAS Source |
|---|---|---|
| ABCA1 | rs1800978 | [63][35] |
| ABCA7 | rs12151021, rs3752231, rs3752246, rs4147929 | [63,64,69,70,71][35][36][37][38][39] |
| ABI3 | rs616338 | [63][35] |
| ACE | rs4277405, rs6504163 | [63,64][35][36] |
| ADAM10 | rs442495, rs593742 | [65,69,71][37][39][40] |
| ADAM17 | rs72777026 | [63][35] |
| ADAMTS1 | rs2830489, rs2830500 | [63,71][35][39] |
| ALPK2 | rs76726049 | [65][40] |
| ANKH | rs112403360 | [63][35] |
| APH1B | rs117618017 | [63,64,65][35][36][40] |
| APOE | rs429358 | [63,64,71][35][36][39] |
| BIN1 | rs4663105, rs6733839 | [63,][35]64,[36]65,[69,70,37][38][39]71[40] |
| BLNK | rs6584063 | [63][35] |
| CASS4 | rs6014724, rs6024870, rs6069737, rs7274581 | [63,64,69,70,71][35][36][37][38][39] |
| CELF1 | rs10838725 | [70][38] |
| CD2AP | rs7767350, rs9369716, rs9381563, rs9473117, rs10948363 | [63,64,65,69,70,71][35][36][37][38][39][40] |
| CD33 | rs1354106, rs3865444, rs12459419 | [64,65,69][36][37][40] |
| CLNK | rs4504245, rs6448453, rs6846529 | [63,64,65][35][36][40] |
| CLU | rs1532278, rs4236673, rs9331896, rs11787077 | [63,64,65,69,70,71][35][36][37][38][39][40] |
| CNTNAP2 | rs114360492 | [65][40] |
| COX7C | rs62374257 | [63][35] |
| CR1 | rs679515, rs2093760, rs4844610, rs6656401 | [63,64,65,69,70,71][35][36][37][38][39][40] |
| CSTF1 | rs6069736 | [69][37] |
| CTSB | rs1065712 | [63][35] |
| CTSH | rs12592898 | [63][35] |
| CYB561 | rs138190086 | [69,71][37][39] |
| DOC2A | rs1140239 | [63][35] |
| ECHDC3 | rs7920721 | [69,71][37][39] |
| EED | rs3851179 | [63,71][35][39] |
| EPHA1 | rs3935067, rs7810606, rs10808026, rs11771145 | [63,64,65,69,70,71][35][36][37][38][39][40] |
| FERMT2 | rs7146179, rs17125924, rs17125944 | [63,64,69,70,71][35][36][37][38][39] |
| FOXF1 | rs16941239 | [63][35] |
| GPR141 | rs2718058 | [70][38] |
| GRN | rs5848 | [63][35] |
| HESX1 | rs184384746 | [65][40] |
| HLA-DQA1 | rs1846190, rs6605556, rs6931277, rs9271192 | [63,64,65,70][35][36][38][40] |
| HLA-DRB1 | rs9271058 | [71][39] |
| ICA1 | rs10952097 | [63][35] |
| IL34 | rs4985556 | [63,69][35][37] |
| INPP5D | rs7597763, rs10933431, rs35349669 | [63,64,65,69,70,71][35][36][37][38][39][40] |
| IQCK | rs7185636 | [71][39] |
| MEF2C | rs190982 | [70][38] |
| MINDY2 | rs602602 | [63,64][35][36] |
| MME | rs16824536, rs61762319 | [63][35] |
| MS4A4A | rs1582763, rs2081545 | [63,64,65,69][35][36][37][40] |
| MS4A6A | rs983392, rs7933202 | [70,[3871]][39] |
| MYO15A | rs2242595 | [63][35] |
| NCK2 | rs115186657, rs143080277 | [64,65][36][40] |
| NECTIN2 | rs41289512 | [65,69][37][40] |
| NYAP1 | rs12539172 | [71][39] |
| OARD1 | rs114812713 | [71][39] |
| PICALM | rs867611, rs561655, rs10792832 | [64,65,69,[37][3870][36]][40] |
| PLCG2 | rs12444183, rs12446759, rs72824905 | [63,69][35][37] |
| PRDM7 | rs56407236 | [63][35] |
| PRKD3 | rs17020490 | [63][35] |
| PSMC3 | rs12292911 | [69][37] |
| PTK2B | rs28834970, rs73223431 | [63,[70,3571]][38][39] |
| RASGEF1C | rs113706587 | [63][35] |
| SCIMP | rs7225151, rs113260531 | [63,65,69][35][37][40] |
| SEC61G | rs76928645 | [63][35] |
| SHARPIN | rs34173062 | [63][35] |
| SLC24A4 | rs7401792, rs10498633, rs12590654, rs12881735 | [63,64,65,69,70,71][35][36][37][38][39][40] |
| SORL1 | rs11218343, rs74685827 | [63,64,65,69,70,71][35][36][37][38][39][40] |
| SORT1 | rs141749679 | [63][35] |
| SPDYE3 | rs7384878 | [63,64][35][36] |
| SPI1 | rs3740688, rs10437655 | [63,64,71][35][36][39] |
| SPPL2A | rs8025980, rs59685680 | [63,69][35][37] |
| TMEM121 | rs7157106, rs10131280 | [63][35] |
| TPCN1 | rs6489896 | [63][35] |
| TREM2 | rs75932628, rs143332484 | [63,71][35][39] |
| TREML2 | rs9381040, rs60755019 | [63,69][35][37] |
| TSPAN14 | rs6586028 | [63][35] |
| UMAD1 | rs6943429 | [63][35] |
| UNC5CL | rs10947943, rs187370608 | [63,64,65][35][36][40] |
| USP6NL | rs7912495, rs11257238 | [63,64,65][35][36][40] |
| WDR12 | rs139643391 | [63][35] |
| WDR81 | rs35048651 | [63][35] |
| WNT3 | rs199515 | [63][35] |
| WWOX | rs62039712 | [71][39] |
| ZCWPW1 | rs1476679 | [69,70][37][38] |
| ZNF652 | rs28394864 | [64,65][36][40] |
References
- Alzheimer’s Disease International. World Alzheimer Report 2018. The State of the Art of Dementia Research: New Frontiers. Available online: https://www.alzint.org/resource/world-alzheimer-report-2018/ (accessed on 15 June 2023).
- GBD 2019 Dementia Forecasting Collaborators. Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: An analysis for the Global Burden of Disease Study 2019. Lancet Public Health 2022, 7, e105–e125.
- Alzheimer’s Association. 2023 Alzheimer’s Disease Facts and Figures. 2023. Available online: https://www.alz.org/media/documents/alzheimers-facts-and-figures.pdf (accessed on 15 June 2023).
- Wimo, A.; Seeher, K.; Cataldi, R.; Cyhlarova, E.; Dielemann, J.L.; Frisell, O.; Guerchet, M.; Jönsson, L.; Malaha, A.K.; Nichols, E.; et al. The worldwide costs of dementia in 2019. Alzheimer’s Dement. 2023, 19, 2865–2873.
- Alzheimer, A. Über Eigenartige Krankheitsfälle des Späteren Alters. Z. Gesamte Neurol. Psychiatr. 1911, 4, 356–385.
- Kokmen, E. The Early Story of Alzheimer’s Disease: Translation of the Historical Papers by Alois Alzheimer, Oskar Fischer, Francesco Bonfiglio, Emil Kraepelin, and Gaetano Perusini. Mayo Clin Proc. 1988, 63, 217–218.
- Grundke-Iqbal, I.; Wisniewski, H.M.; Johnson, A.B.; Terry, R.D.; Iqbal, K. Evidence that Alzheimer neurofibrillary tangles originate from neurotubules. Lancet 1979, 1, 578–580.
- Masters, C.L.; Simms, G.; Weinman, N.A.; Multhaup, G.; McDonald, B.L.; Beyreuther, K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. USA 1985, 82, 4245–4249.
- Kosik, K.S.; Joachim, C.L.; Selkoe, D.J. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. USA 1986, 83, 4044–4048.
- Braak, H.; Braak, E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82, 239–259.
- Ball, M.; Braak, H.; Coleman, P.; Dickson, D.; Duyckaerts, C.; Gambetti, P.; Hansen, L.; Hyman, B.; Jellinger, K.; Markesbery, W.; et al. Consensus Recommendations for the Postmortem Diagnosis of Alzheimer’s Disease. Neurobiol. Aging 1997, 18, S1–S2.
- Kim, C.K.; Lee, Y.R.; Ong, L.; Gold, M.; Kalali, A.; Sarkar, J. Alzheimer’s Disease: Key Insights from Two Decades of Clinical Trial Failures. J. Alzheimer’s Dis. 2022, 87, 83–100.
- Kehoe, P.G.; Turner, N.; Howden, B.; Jarutyte, L.; Clegg, S.L.; Malone, I.B.; Barnes, J.; Nielsen, C.; Sudre, C.H.; Wilson, A.; et al. Safety and efficacy of losartan for the reduction of brain atrophy in clinically diagnosed Alzheimer’s disease (the RADAR trial): A double-blind, randomised, placebo-controlled, phase 2 trial. Lancet Neurol. 2021, 20, 895–906.
- Haeberlein, S.B.; Aisen, P.; Barkhof, F.; Chalkias, S.; Chen, T.; Cohen, S.; Dent, G.; Hansson, O.; Harrison, K.; von Hehn, C.; et al. Two Randomized Phase 3 Studies of Aducanumab in Early Alzheimer’s Disease. J. Prev. Alzheimer’s Dis. 2022, 9, 197–210.
- Knopman, D.S.; Jones, D.T.; Greicius, M.D. Failure to demonstrate efficacy of aducanumab: An analysis of the EMERGE and ENGAGE trials as reported by Biogen, December 2019. Alzheimer’s Dement. 2021, 17, 696–701.
- Alexander, G.C.; Emerson, S.; Kesselheim, A.S. Evaluation of Aducanumab for Alzheimer Disease: Scientific Evidence and Regulatory Review Involving Efficacy, Safety, and Futility. JAMA 2021, 325, 1717–1718.
- Dunn, B.; Stein, P.; Cavazzoni, P. Approval of Aducanumab for Alzheimer Disease—The FDA’s Perspective. JAMA Intern. Med. 2021, 181, 1276–1278.
- McDade, E.; Cummings, J.L.; Dhadda, S.; Swanson, C.J.; Reyderman, L.; Kanekiyo, M.; Koyama, A.; Irizarry, M.; Kramer, L.D.; Bateman, R.J. Lecanemab in patients with early Alzheimer’s disease: Detailed results on biomarker, cognitive, and clinical effects from the randomized and open-label extension of the phase 2 proof-of-concept study. Alzheimer’s Res. Ther. 2022, 14, 191.
- Van Dyck, C.H.; Swanson, C.J.; Aisen, P.; Bateman, R.J.; Chen, C.; Gee, M.; Kanekiyo, M.; Li, D.; Reyderman, L.; Cohen, S.; et al. Lecanemab in Early Alzheimer’s Disease. N. Engl. J. Med. 2023, 388, 142–143.
- Xu, R.; Li, X.; Boreland, A.J.; Posyton, A.; Kwan, K.; Hart, R.P. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 2020, 11, 1577.
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.; Bohlen, C.; Adil, A.; Tucker, A.; et al. New tools for studying microglia in the mouse human, C.N.S. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746.
- Satoh, J.-I.; Kino, Y.; Asahina, N.; Takitani, M.; Miyoshi, J.; Ishida, T.; Saito, Y. TMEM119 marks a subset of microglia in the human brain. Neuropathology 2016, 36, 39–49.
- Vankriekelsvenne, E.; Chrzanowski, U.; Manzhula, K.; Greiner, T.; Wree, A.; Hawlitschka, A.; Llovera, G.; Zhan, J.; Joost, S.; Schmitz, C.; et al. Transmembrane protein 119 is neither a specific nor a reliable marker for microglia. Glia 2022, 70, 1170–1190.
- Green, K.N.; Crapser, J.D.; Hohsfield, L.A. To Kill a Microglia: A Case for CSF1R Inhibitors. Trends Immunol. 2020, 41, 771–784.
- Spangenberg, E.; Severson, P.L.; Hohsfield, L.A.; Crapser, J.; Zhang, J.; Burton, E.A.; Zhang, Y.; Spevak, W.; Lin, J.; Phan, N.Y.; et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat. Commun. 2019, 10, 3758.
- Muffat, J.; Li, Y.; Yuan, B.; Mitalipova, M.; Omer, A.; Corcoran, S.; Bakiasi, G.; Tsai, L.-H.; Aubourg, P.; Ransohoff, R.M.; et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016, 22, 1358–1367.
- Dräger, N.M.; Sattler, S.M.; Huang, C.T.-L.; Teter, O.M.; Leng, K.; Hashemi, S.H.; Hong, J.; Aviles, G.; Clelland, C.D.; Zhan, L.; et al. A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat. Neurosci. 2022, 25, 1149–1162.
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293.e9.
- Pandya, H.; Shen, M.J.; Ichikawa, D.M.; Sedlock, A.B.; Choi, Y.; Johnson, K.R.; Kim, G.; Brown, M.A.; Elkhaloun, A.G.; Maric, D.; et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 2017, 20, 753–759.
- Chen, S.-W.; Hung, Y.-S.; Fuh, J.-L.; Chen, N.-J.; Chu, Y.-S.; Chen, S.-C.; Wong, Y.-H. Stem Cell Reports Resource Efficient conversion of human induced pluripotent stem cells into microglia by defined transcription factors. Stem Cell Rep. 2021, 16, 1363–1380.
- Brownjohn, P.W.; Smith, J.; Solanki, R.; Lohmann, E.; Houlden, H.; Hardy, J.; Dietmann, S.; Livesey, F.J. Functional Studies of Missense TREM2 Mutations in Human Stem Cell-Derived Microglia. Stem Cell Rep. 2018, 10, 1294–1307.
- Douvaras, P.; Sun, B.; Wang, M.; Kruglikov, I.; Lallos, G.; Zimmer, M.; Terrenoire, C.; Zhang, B.; Gandy, S.; Schadt, E.; et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Rep. 2017, 8, 1516–1524.
- Speicher, A.M.; Wiendl, H.; Meuth, S.G.; Pawlowski, M. Generating microglia from human pluripotent stem cells: Novel in vitro models for the study of neurodegeneration. Mol. Neurodegener. 2019, 14, 46.
- Andrews, S.J.; Renton, A.E.; Fulton-Howard, B.; Podlesny-Drabiniok, A.; Marcora, E.; Goate, A.M. The complex genetic architecture of Alzheimer’s disease: Novel insights and future directions. EBioMedicine 2023, 90, 104511.
- Bellenguez, C.; Küçükali, F.; Jansen, I.E.; Kleineidam, L.; Moreno-Grau, S.; Amin, N.; Naj, A.C.; Campos-Martin, R.; Grenier-Boley, B.; Andrade, V.; et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 2022, 54, 412–436.
- Wightman, D.P.; Jansen, I.E.; Savage, J.E.; Shadrin, A.A.; Bahrami, S.; Holland, D.; Rongve, A.; Børte, S.; Winsvold, B.S.; Drange, O.K.; et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 2021, 53, 1276–1282.
- Marioni, R.E.; Harris, S.E.; Zhang, Q.; McRae, A.F.; Hagenaars, S.P.; Hill, W.D.; Davies, G.; Ritchie, C.W.; Gale, C.R.; Starr, J.M.; et al. GWAS on family history of Alzheimer’s disease. Transl. Psychiatry 2018, 8, 99.
- Lambert, J.-C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458.
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Olaso, R.; Garnier, J.G.; Moutet, M.L.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430.
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413.
- Dardiotis, E.; Siokas, V.; Pantazi, E.; Dardioti, M.; Rikos, D.; Xiromerisiou, G.; Markou, A.; Papadimitriou, D.; Speletas, M.; Hadjigeorgiou, G.M. A novel mutation in TREM2 gene causing Nasu-Hakola disease and review of the literature. Neurobiol. Aging 2017, 53, 194.e13–194.e22.
- Velasco, S.; Kedaigle, A.J.; Simmons, S.K.; Nash, A.; Rocha, M.; Quadrato, G.; Paulsen, B.; Nguyen, L.; Adiconis, X.; Regev, A.; et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 2019, 570, 523–527.
- Lancaster, M.A.; Corsini, N.S.; Wolfinger, S.; Gustafson, E.H.; Phillips, A.W.; Burkard, T.R.; Otani, T.; Livesey, F.J.; Knoblich, J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017, 35, 659–666.
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Bräuninger, M.; Lewitus, E.; Sykers, A.; Hevers, W.; Lancaster, M.; et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677.
- Song, L.; Yuan, X.; Jones, Z.; Vied, C.; Miao, Y.; Marzano, M.; Hua, T.; Sang, Q.-X.A.; Guan, J.; Ma, T.; et al. Functionalization of Brain Region-specific Spheroids with Isogenic Microglia-like Cells. Sci. Rep. 2019, 9, 11055.
- Andrews, M.G.; Kriegstein, A.R. Challenges of organoid research. Annu. Rev. Neurosci. 2022, 45, 23–39.
- Quadrato, G.; Brown, J.; Arlotta, P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat. Med. 2016, 22, 1220–1228.
- Uzquiano, A.; Kedaigle, A.J.; Pigoni, M.; Paulsen, B.; Adiconis, X.; Kim, K.; Faits, T.; Nagaraja, S.; Antón-Bolaños, N.; Gerhardinger, C.; et al. Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell 2022, 185, 3770–3788.e27.
